2025 AIChE Annual Meeting
(73d) Tunable Self-Assembling Single-Neutrophil Extracellular Trap Device
Authors
Another neutrophil function, which is not a multicellular response, is NETosis, a neutrophil-specific type of cell death where traps are released into the extracellular space containing histones, chromatin, and/or mitochondrial DNA. The specific type of NETosis determines what is released. Suicidal NETosis involves the complete decondensation of the nucleus, spewing histones, chromatin, neutrophil elastase (NE) and myeloperoxidase (MPO) around itself and at its target, resulting in cell death. In contrast, vital NETosis has two independent mechanisms: one where PAD4 is activated and chromatin is decondensed and packaged into vesicles along with proteins used in pathogen elimination (e.g., MPO and NE), and another where mitochondrial DNA is packaged into vesicles and released (Figure 1A). These neutrophils remain vital, maintaining the ability to phagocytose and respond to chemotaxis.
Many engineered techniques have been developed to study the diverse functions of neutrophils. These include patterning clusters of BioparticlesTM to initiate neutrophil swarming with microscopic analysis, designing PDMS chambers to enclose 1-4 neutrophils and capture released NETs, and creating devices to capture NETs themselves to study their effects on different bacteria. However, none of these techniques consistently achieve single-cell resolution, and they only study the effects of suicidal NETosis.
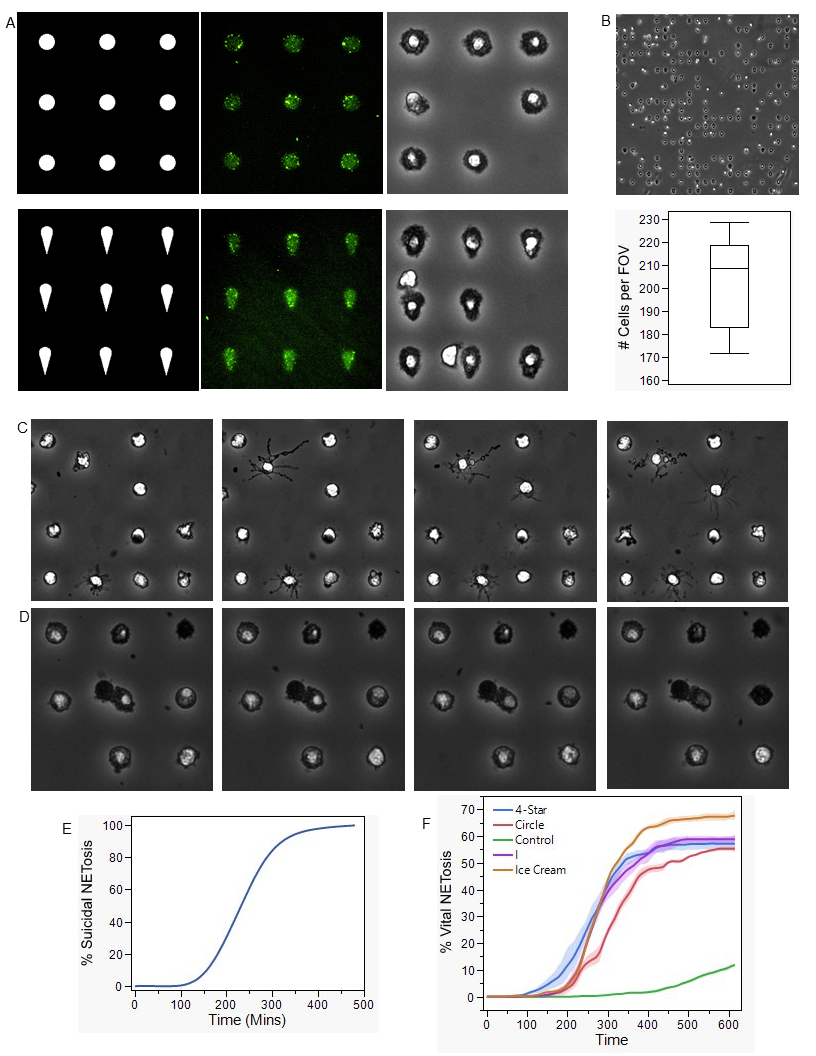
Herein, we present a tunable, self-assembling single neutrophil NETosis device. Neutrophils are consistently patterned down to single-cell resolution along a grid of bacterial outer membrane vesicles (OMVs), allowing the study of both suicidal and vital NETosis within the same device, depending on the desired outcome. Through light-induced extracellular vesicle adsorption, OMVs are patterned along a uniform 100x100 array of 16π μm² area shapes, onto which neutrophils self-assemble onto the array and undergo NETosis.
Methods: pET/EGFP-strep-his plasmids were used to produce the GFP OMVs. The plasmids were miniprepped according to the manufacturer's protocol (Qiagen) and transformed into competent E. coli BLR (DE3) cells. After growth, the culture was centrifuged at 8,000 x g to pellet the whole cells and large vesicles, leaving the OMVs in the supernatant. This supernatant was sterile vacuum-filtered through a 0.2 µm filter. The OMV media was further enriched and purified using tangential flow filtration with a molecular weight cut-off (MWCO) of 500 kDa using polysulfone hollow fiber filter cartridges with seven dia-cycles. The enriched media was then further concentrated using a 50 kDa spin filter (Amicon) at 3,000 x g until the desired volume was achieved. The resulting OMVs were stored at –80°C until use. All OMVs were characterized using tunable resistive pulse sensing (TRPS) and transmission electron microscopy (TEM).
Human peripheral blood was collected in K2-Ethylenediaminetetraacetic acid (EDTA), purple top, tubes. Fresh blood is added to an RBC lysis buffer solution and is centrifuged to isolate the white blood cells, while the supernatant is discarded. The neutrophils are isolated using immuno-magnetic negative selection with the EasySepTM Human Neutrophil Isolation Kit according to the manufacturers protocol. The neutrophils were stained in serum free Iscove’s Modified Dulbecco’s Medium (IMDM) and 1% penicillin-streptomycin (PS) with 1nM of CellTracker Green and 20 µg/mL Hoechst 33342. The final neutrophil suspension is at 1.2 x 106 cells / mL in IMDM with 20% fetal bovine serum and 1% PS.
Initially, glass coverslips were cycled between ethanol and deionized (DI) water for 3 sonication cycles to ensure cleanliness. The surface of the coverslip was then activated via oxygen plasma for 1 minute and a silicon well was placed atop the glass. Into the wells a 0.01% (w/v) poly-L-lysine (PLL) was incubated on the surface for 1 hour. Following 3 washings of 0.1M caustic HEPES buffer the surface is incubated with 100 mg/mL methoxy-poly(ethylene glycol)-succinimidyl valerate (mPEG-SVA) diluted in 0.1M HEPES buffer for 1 hour. The surface is rinsed in DI water and dried under nitrogen flow before a benzophenone based photoactivator, 4-benzoylbenzyl-trimethylammonium chloride (PLPP), diluted in 96% Ethanol was placed onto the surface through evaporative deposition. The coverslips were photoetched utilizing a digital micro-mirror optical module (PRIMO) mounted on an automated inverted microscope, with a resolution down to 1µm. The UV does was 30mJ/mm2. The patterns were designed with the microscope camera resolution of 0.29µm / pixel for area calculations. The remaining PLPP is washed away with DI water and the surface is dried under nitrogen flow. The surface is rehydrated with phosphate buffer solution (PBS) for 5 minutes followed by E11 OMVs being added to adsorb to the photoetched regions, followed by 10 washes of PBS. The surface is coated with FBS for 5 minutes to aid in neutrophil adhesion.
50µL of final neutrophil solution was added to a well and the cells were allowed to settle on the patterned OMV surface to pattern themselves for 1 hour at 37C. For no PMA addition, the wells are sealed with a 12mm round coverslip and imaged in a Nikon Ti2 equipped with Ozark stage top incubator every 10 minutes for 7 hours. For PMA addition, the supernant is gently removed and replaced with media spiked with 4nM PMA. This sample is then imaged in the same manner as above.
Results: To produce the stimuli needed for NETosis induction, E. coli pET/eGFP-strept-his plasmid was transformed into competent BLR(DE3) cells, allowing them to express eGFP to ensure encapsulation of the protein into the OMVs. OMVs are known to be most produced during cell division; therefore, induction was targeted to the late log phase for optimal OMV production. To separate the OMVs from the broth containing excess proteins, tangential flow filtration (TFF) was utilized. In this process, the OMVs are retained while the unwanted proteins are diluted out through continuous diafiltration with PBS. This method has been validated to produce extracellular particles of increasing purity. To quantify OMV concentration and size distribution, tunable resistive pulse sensing was utilized. A TEM image of purified OMVs validates this size distribution.
Four different patterns were engineered to be utilized by the digital micromirror device within PRIMOTM to stimulate the neutrophils in different ways. The circular pattern was designed to maintain uniformity of the cell. The ice cream cone pattern was designed as an asymmetric pattern to potentially increase the amount of NETosis (Figure A). At a resolution down to 1 µm, OMVs can adhere to the patterned regions which were exposed to UV light (Figure A). Neutrophils will take the shape of the desired patterns, indicating that the design is affecting the neutrophils' stimulation (Figure A). This effect can be seen in a wide field of view of a 23x23 array of OMV patterns, showcasing the expandability of this platform (Figure B). The average number of cells per FOV is 210 when imaged under a 20x objective (Figure B).
Neutrophil patterning with OMVs can be utilized to induce vital NETosis as seen through the vesiculation and protrusions coming from the neutrophil’s membrane (Figure C). Where the ice cream patterned OMVs induce NETosis in up to 70%, which is significantly different than the other patterns tested (Figure F). When PMA is added to induce suicidal NETosis 100% of the cells undergo suicidal NETosis (Figure E). Neutrophil will appear as if they are dying when undergoing suicidal NETosis (Figure D).
Implications: Herein, we present a novel neutrophil extracellular trap device which can measure either suicidal or vital NETosis depending on the desired outcome at a single cell level. This device is the first of its kind to be able to do this and this opens many new avenues to explore outside effects on NETosis. There is currently no method out there to be able to measure vital NETosis, therefore its implication on many diseases is still unknown, with this device it opens up the ability to deeply understand the mechanisms of vital NETosis and the expandability of the platform allows many conditions to be running in parallel, allowing the user to test and preform many desired outcomes.