2025 AIChE Annual Meeting
(415i) Size- and Charge-Dependent Microrheology in Live Escherichia coli: Impact of Confinement and Macromolecular Interactions on Particle Dynamics and Localization
Authors
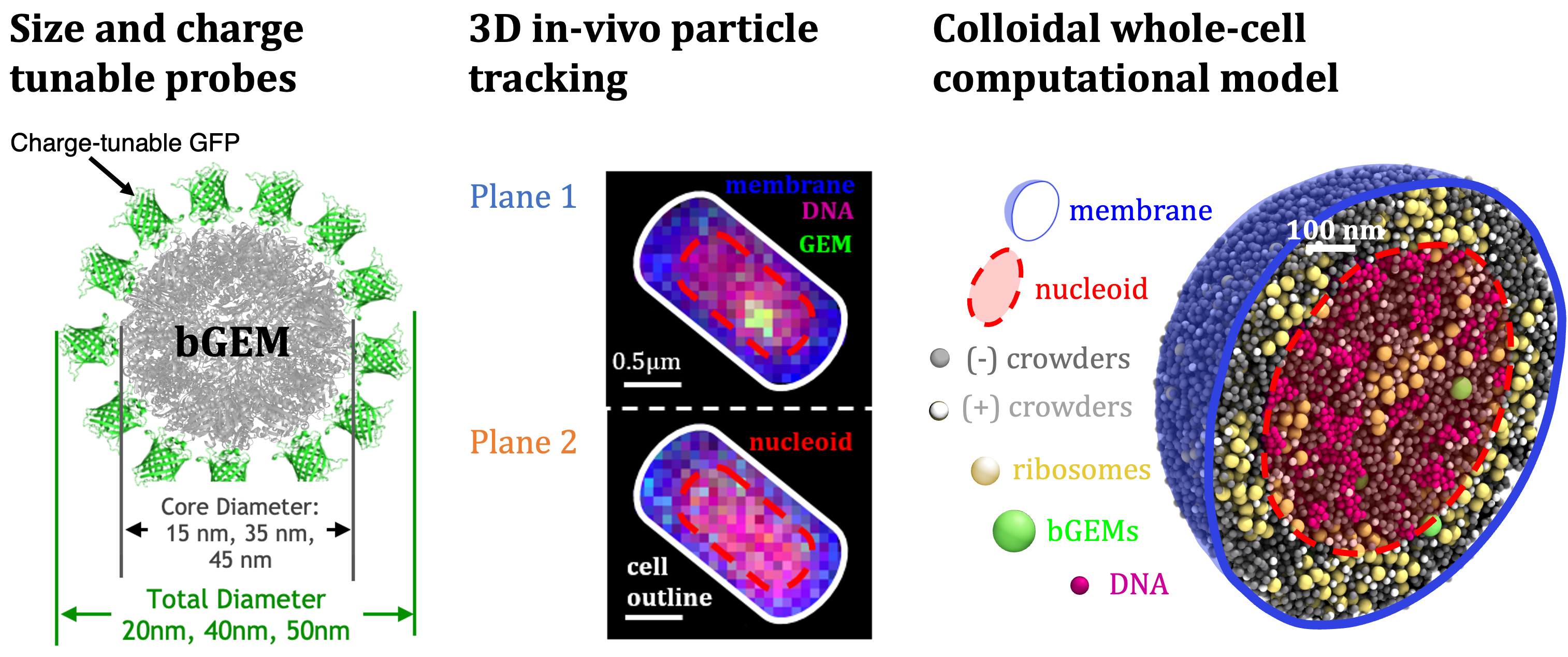
We identify entropic (size-based) and electrostatic (charge-based) mechanisms that drive spatial segregation, finding smaller (20 nm) particles enriched within the nucleoid, while larger or positively charged particles are excluded. This localization emerges from the interplay of cytoplasmic polydispersity, nucleoid architecture, and interactions with cellular components such as ribosomes and DNA. Critically, we show that previously reported subdiffusive particle dynamics are primarily due to geometrical confinement rather than intrinsic anomalous diffusion. Although single-molecule tracking at moderate temporal resolutions suggests universal subdiffusion, simulations with higher temporal resolution clarify that confinement within the nucleoid and cell boundaries governs this apparent anomalous diffusion. Thus, our results provide new insight into intracellular transport, highlighting the crucial roles of confinement and macromolecular crowding in biomolecular localization and diffusion. More broadly, this work exemplifies how explicit modeling of cell-spanning biomolecular dynamics with high-performance simulations enables the discovery of emergent behaviors driven by biophysical and biochemical interactions – laying the groundwork for uncovering new molecular mechanisms, developing physics-based digital twins, and engineering cells for applications such as energy generation and disease treatment.