2025 AIChE Annual Meeting
(686g) Optogenetic Control of BMP Signaling Dynamics in Cells and Tissues
Dynamic signaling pathways regulate critical cell fate decisions during development and tissue maintenance. Among these, the Bone Morphogenetic Protein (BMP) pathway plays a conserved role in early development, establishing spatial patterning, and maintaining stem cells across animal systems. Yet, the real-time dynamics of BMP signaling and how cells decode these signals remain poorly understood. To quantitatively understand how extracellular signals are processed into specific intracellular responses, we have developed an integrated live-cell platform to monitor and precisely manipulate BMP pathway activity in Drosophila melanogaster using optogenetics and fluorescent reporters.
Our platform combines genetically encoded fluorescent biosensors with optogenetic actuators to dissect BMP signaling at cellular and tissue levels. Specifically, we use fluorescently tagged versions of the signaling co-factor Smad4 (Drosophila homolog: Medea) to visualize nuclear translocation and DNA-binding behavior in real time (Fig. 1). To manipulate pathway activation, we engineered optogenetic BMP receptors by fusing the Light-Oxygen-Voltage (LOV) domain to the Type I and Type II BMP receptors, Thickveins (Tkv) and Punt. This enables blue light-induced receptor dimerization and pathway activation (Fig. 3). On the other hand, the Cryptochrome 2 (Cry2) system allows for light-induced clustering of signaling components to inhibit BMP transduction with precise timing (Fig. 4).
These tools were first validated in Drosophila-derived Schneider 2 (S2) cells, which provide a controlled background for BMP studies due to the absence of endogenous ligands. To extend this work into in vivo Drosophila tissues, we are generating and validating transgenic fly lines expressing opto-receptors and Medea (Med) fused to various fluorescent proteins (GFP, mScarlet-I, mTagBFP2, smURFP) under UAS promoter for tissue-specific visualization. In parallel, a CRISPR-based Med-GFP knock-in line is being developed to endogenously report Med dynamics in embryos and germline stem cells (GSCs). These tools will be used to analyze Med responses in the embryonic BMP gradient and in the GSC niche, where BMP signaling regulates asymmetric division and stem cell maintenance (Fig. 2).
Altogether, our work establishes a modular and generalizable framework for probing in vivo morphogen signaling dynamics in live multicellular systems. The combination of optogenetic control and live dynamic imaging enables deeper insight into how cells interpret signals over time and space, with potential applications in developmental biology, regenerative medicine, and synthetic biology.
Figures:
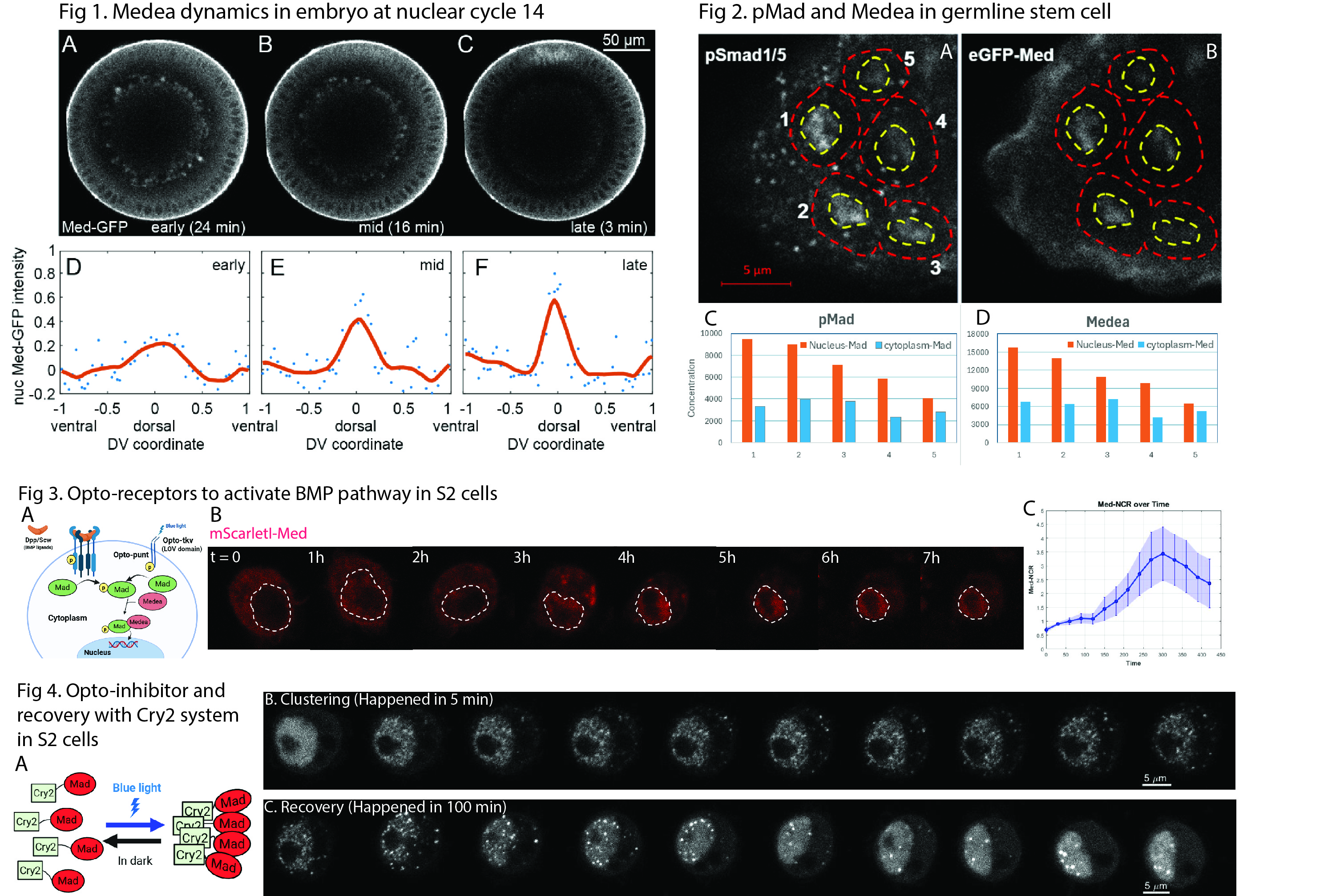
Fig. 1. Cross-sectional expression and intensity profiles of GFP-Medea along the dorsal–ventral axis in Drosophila embryos at nuclear cycle 14: (A, D) early stage, (B, E) mid stage, and (C, F) late stage.
Fig. 2. Immunostaining of pSmad1/5 and Medea in the Drosophila germarium. In panels A and B, cells labeled 1 and 2 are germline stem cells (GSCs), cell 3 is a pre-cystoblast, and cells 4 and 5 are cystoblasts. The concentration gradients of both proteins across these cells are shown in charts C and D.
Fig. 3. (A) Schematic representation of optogenetic BMP receptors, which fuse the LOV domain with BMP receptors to enable pathway activation by blue light. (B) Activation process showing Medea translocation from the cytoplasm to the nucleus. (C) Intensity curve of nuclear localization over time.
Fig. 4. (A) Schematic of the Cry2 optogenetic system. (B) Light-induced clustering process activated by blue light. (C) Recovery process following removal of stimulation.