2025 AIChE Annual Meeting
(271b) Native Single-Cell Western Blot: Measuring Heterogeneity of Multicomponent Protein Complexes in Cancer Cell Populations
Protein-protein interactions (PPIs) play crucial roles in cell homeostasis, including molecular pathways, gene regulation, signaling, and more. Changes associated to the binding kinetics, abundance, or subunit composition of multimeric protein complexes are linked to disease. Particularly in cancer, chaperone proteins form stable multicomponent complexes and have been suggested as a survival mechanism in this stressed cell population1. Often referred to as the ‘epichaperome’, this stable multimeric network could point to novel therapeutic avenues that target aberrant network interactions as opposed to previously tested individual chaperone inhibition1,2.
Measurement of multimeric complexes in cancer cell populations requires novel single-cell analysis approaches to fully assess their inherent heterogeneity, however, current technologies are limited to pair interactions (FRET, PLA) or lack the necessary throughput to identify subpopulations. Microfluidic technologies are widely employed for single-cell analysis and established protocols have been previously used for measuring small, denatured proteins and monomeric complexes from single cells3,4. Still, challenges remain in measurements of multimeric protein complexes as binding kinetics and diffusive losses can reduce assay sensitivity. To address these limitations, we are developing a native single-cell western blot platform to spatially separate protein complexes and measure their abundance from 100s-1000s of individual cells, while preserving multimeric interactions.
As a case study, we show how native single-cell western blot can be used to measure chaperone-associated complexes from the pediatric cancer Ewing Sarcoma. This disease is characterized for its fusion oncoprotein driver EWS-FLI1, an intrinsically disordered protein known for interacting with the abundant chaperone HSP90. Changes in the expression levels of EWS-FLI1 have been linked to cellular phenotype changes leading to migratory or proliferative phenotypes5. Therefore, measuring changes in chaperone networks in populations with altered levels of EWS-FLI1 could point to new therapeutic strategies and better understanding of chaperone PPIs in disease.
Methods:
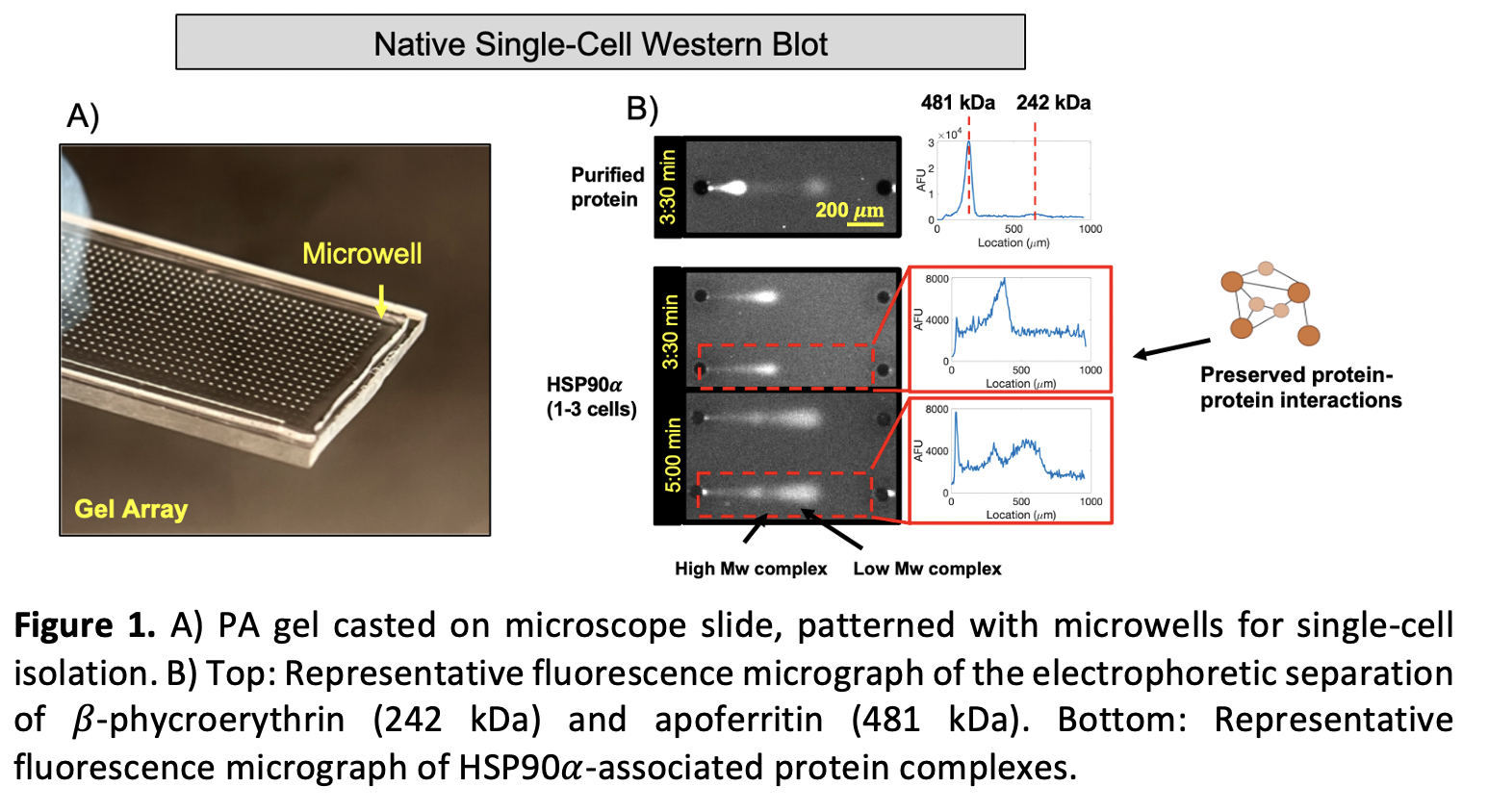
We miniaturize traditional electrophoretic separations to be able to measure endogenous protein complexes from individual cancer cells. To do this, standard photolithography methods are employed to obtain arrays of SU-8 cylindrical microposts on silicon wafers, which are then used to pattern a thin (30 – 40 µm) polyacrylamide (PA) layer on a microscope slide6. The post dimensions are carefully tuned to isolate individual cells in the resulting microwells (Figure 1 A)).
Individual cells are gravity-settled into the microwells of the gel array, which is then placed inside of a custom 3D-printed electrophoresis and flow chamber. Precise control of the buffer temperature and flow rate decrease Joule heating and local pH gradients that can cause analyte diffusive losses or inconsistent migration patterns of the protein bands. A dual-purpose lysis and electrophoresis buffer is used to quickly lyse cells and run the assay limiting protein complex dissociation. A photoactive benzophenone group is used to covalently link proteins to the PA gel and stop further diffusion of the bands. Lastly, rounds of in gel immunoprobing with fluorescent antibodies are performed to target the proteins of interest6.
The fluorescence intensity and migration distance of each band is quantified using a high-throughput electrophoresis analysis algorithm7 to assess heterogeneity and changes to the molecular weights associated to large protein complexes within cell populations, particularly relevant in highly heterogenous ones, such as cancer.
Results:
We evaluated electromigration of a purified and fluorescently labeled large protein complex (β-phycroerythrin, 242 kDa) with cold dual-purpose lysis and electrophoresis buffer under no-flow and continuous flow conditions. With no flow, a band width of 210.6 µm (CV = 27.5%) and an average migration distance of 604.0 µm (CV = 13.7%) was observed (n = 816). Under flow conditions, band width and migration distance were 73.8 µm (CV = 24.4%) and 565.2 µm (CV = 8.0%), respectively (n = 1003). Overall, the use of a continuous flow of cold buffer aided in a statistically significant reduction of diffusive spreading of the bands (one tailed F-test = 10.35, p<0.001), facilitating their analysis.
Two model multimeric protein complexes of different molecular weights were used to validate the complex separation performance of the assay: β-phycroerythrin (242 kDa) and apoferritin (481 kDa). With an electrophoresis time of 3:30 min and an electric field of 33 V/cm, a separation resolution (Rs) of 2.95 ± 0.95 (n = 332 separations) was obtained, achieving peak baseline resolution (>1.5)(Figure 1B), top).
We then selected the Ewing Sarcoma cell line TC-71 and probed for the chaperone HSP90α, which is responsible for nucleating aberrant chaperone complexes in epichaperomes2. Using a 5 min electrophoresis time, 8%T gel density, and 1-mm separation lanes, we observe the presence of two distinct bands, likely associated to complexes of different molecular weights (Figure 1B), bottom). Previous works have reported the presence of HSP90 complexes in cancer and pluripotent stem cells, appearing as different molecular weight bands in bulk native western blots, linking those to the dimeric form of HSP90 and higher molecular structures composed of other chaperones, such as HOP and HSP702,8. Additional testing is still required to fully assess the molecular weights and identity of the subunits associated to HSP90 bands in the individual Ewing Sarcoma cells analyzed here.
Implications:
Technologies that allow measurement of multicomponent protein complexes linked to disease have not been adapted so far to address the heterogeneity of cell populations. Single-cell analyses based on genomics and transcriptomics do not accurately predict PPIs and protein-based analyses lack the necessary throughput. The method presented in this work aims to address this need by allowing the measurement of endogenous protein complexes from 100s-1000s of cells, to be able to discern possible subpopulations associated with modified protein networks in disease. In Ewing Sarcoma, understanding PPIs of chaperone proteins that interact with oncogenic drivers could inform the next generation of network targeting therapies.
References:
1. Rodina et al. Nature 538 (7625) (2016)
2. Roychowdhury et al. Nat Commun (2024)
3. Hughes et al. Methods11, 749 (2014)
4. Vlassakis & Hansen et al. Nat Commun 12, (2021)
5. Apfelbaum et al. Oncol. (2022)
6. Kang et al. Protoc. 11, (2016)
7. Vlassakis, J. et al. SLAS Technol 26, 637–649 (2021)