2025 AIChE Annual Meeting
(715g) A Multi-Dimensional Feature-Based Workflow for Accelerating Enzyme Discovery
Authors
Xuwei Ding, East China University of Science and Technology
Gaowei Zheng, East China University of Science and Technology
Zheng Liu, Tsinghua University
Recent advancements in data technology have unlocked significant opportunities for the discovery and development of new enzymes for the green synthesis of chemicals. However, current protein databases predominantly focus on overall sequence matches, while the multi-scale features underlying catalytic mechanisms and processes remain scattered across various data sources and insufficiently integrated for effective enzyme mining.
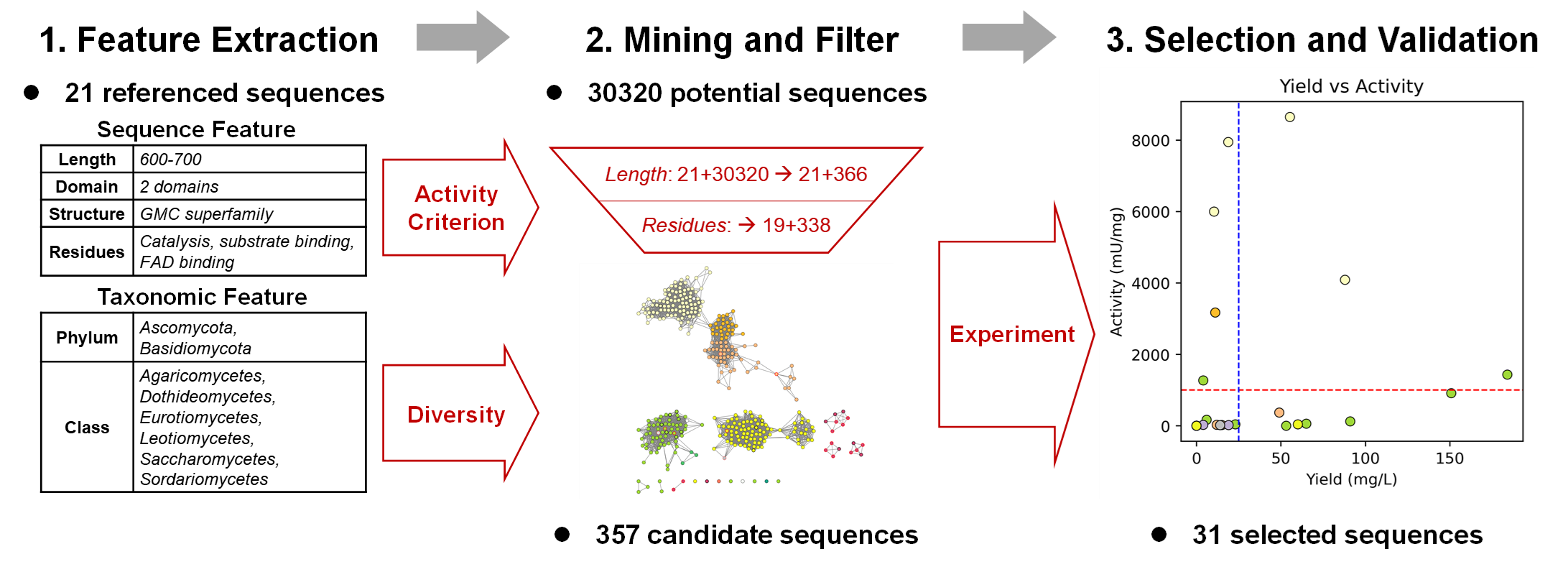
In this study, we developed a workflow driven by sequence and taxonomic feature evaluation to discover enzymes expressed in E. coli and catalyzing chemical reactions in vitro, using alcohol oxidase (AOX) as a demonstration. AOX catalyzes the conversion of methanol to formaldehyde. Using a dataset of 21 reported AOXs, we constructed sequence scoring rules based on features such as sequence length, structural motifs, catalytic residues, binding residues, and overall structure. These scoring rules were applied to refine results from HMM-based searches, yielding 357 candidate sequences of eukaryotic origin, which were categorized into six classes at 85% sequence similarity. Experimental validation was conducted in two rounds on 31 selected sequences representing all classes. Among these, 19 (61.3%) were expressed as soluble proteins in E. coli, and 18 (58.1%) exhibited AOX activity, as predicted—significantly higher than the activity probability of UniProt-based annotation without functional feature evaluation (2/12, 16.7%). Notably, the most active recombinant AOX demonstrated an activity of 8.65 ± 0.29 U/mg, approaching the highest activity of native eukaryotic enzymes. Compared to our previous directed evolution work on AOX, this workflow tested only one-third the number of sequences but yielded enzymes with twice the activity. Furthermore, systematic mining revealed clusters within natural enzyme sequences associated with high activity and expression, providing a good starting point for further enzyme engineering efforts.