2025 AIChE Annual Meeting
(715f) Leveraging Large Language Models for Enzyme Catalytic Efficiency Prediction
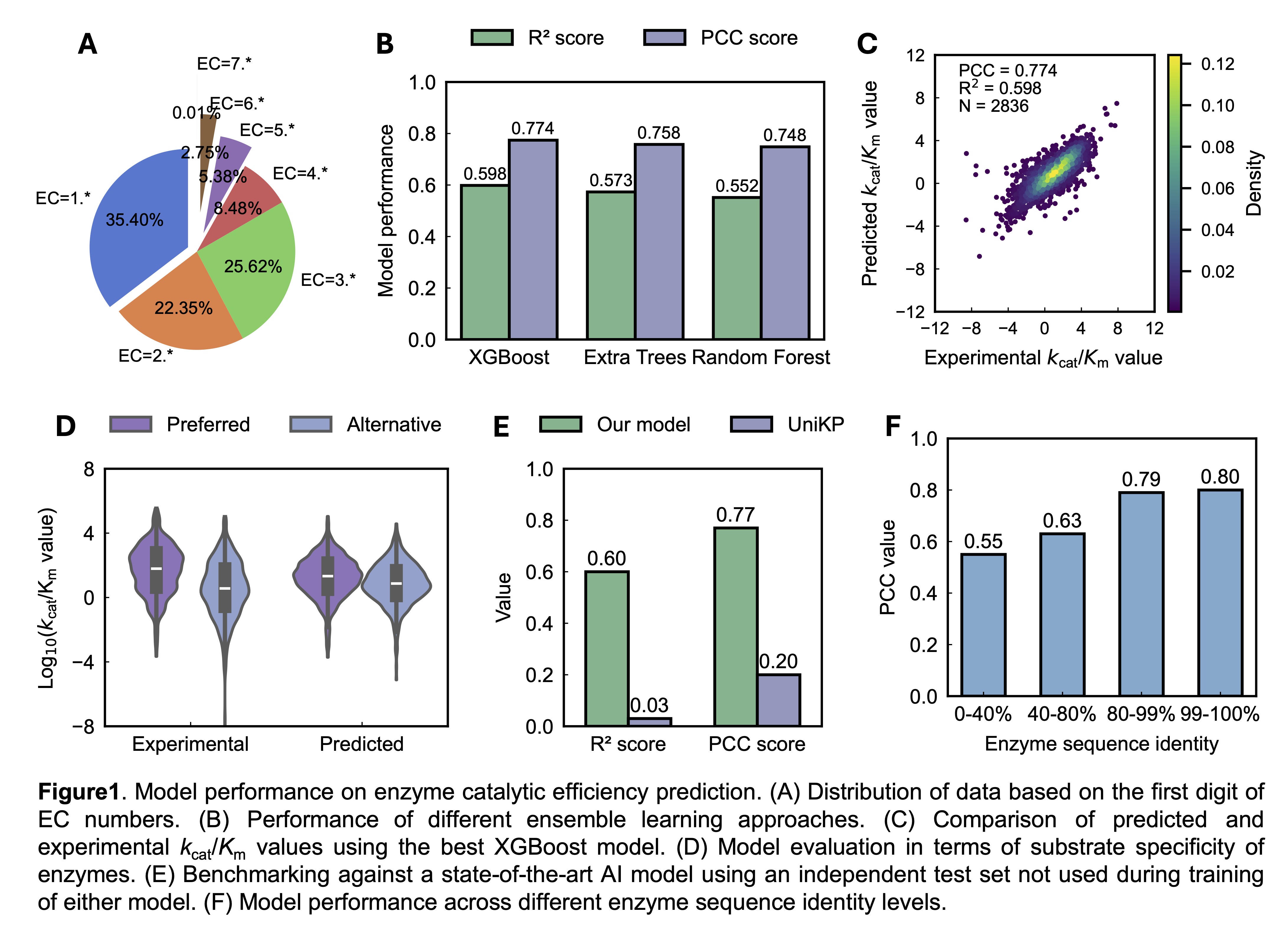
Enzyme catalytic efficiency (kcat/Km) is a fundamental parameter in enzymology, reflecting how effectively an enzyme converts a substrate into a product. It also allows comparison between different enzymes or enzyme variants to determine which is the more efficient under similar conditions. However, experimental determination of kcat/Km values is labor-intensive, time-consuming, and often limited in scale. In this study, we leveraged large language models (LLMs) to predict kcat/Km values directly from enzyme sequences and substrate structures. We first carefully curated a high-quality experimental dataset from open-source databases, encompassing 28,356 unique entries catalyzed by 11,997 unique sequences from 1,560 organisms (Figure 1A). For representations, we utilized advanced protein LLMs (ESM v2, ESM3, and ProtT5) and compound LLMs (ChemBERTa-MTR and ChemBERTa-MLM) to generate embeddings for enzymes and substrates, respectively. These embeddings were concatenated and fed into various ensemble learning models, resulting in excellent predictive performance on the held-out test set (Figure 1B&C). Beyond numerical accuracy, the model effectively captured enzyme substrate preferences. In particular, it distinguished preferred substrates from less favored alternatives, demonstrating strong potential for applications in predicting biological substrate specificity (Figure 1D). When benchmarked against UniKP, a state-of-the-art AI tool for enzyme kinetics prediction, our model achieved significantly better performance on an independent test dataset excluded from the training of both models (Figure 1E). Notably, the model could be generalized even for enzymes with low sequence similarity, highlighting its potential for broad applicability (Figure 1F). We further extended our approach to train a model for the Michaelis constant (Km) using a separate high-quality dataset of over 60,700 unique entries, again achieving great performance. Finally, we applied the trained model to perform genome-scale predictions across 332 yeast species, thus providing a valuable resource for the yeast research community. In conclusion, our findings demonstrate that we developed a powerful LLM-based tool for predicting enzyme catalytic efficiency, with strong implications for enzyme engineering and synthetic biology.