2025 AIChE Annual Meeting
(654g) Invited Talk: Programmable RNA Editing Via a DNA-Guided Cas12a System
Authors
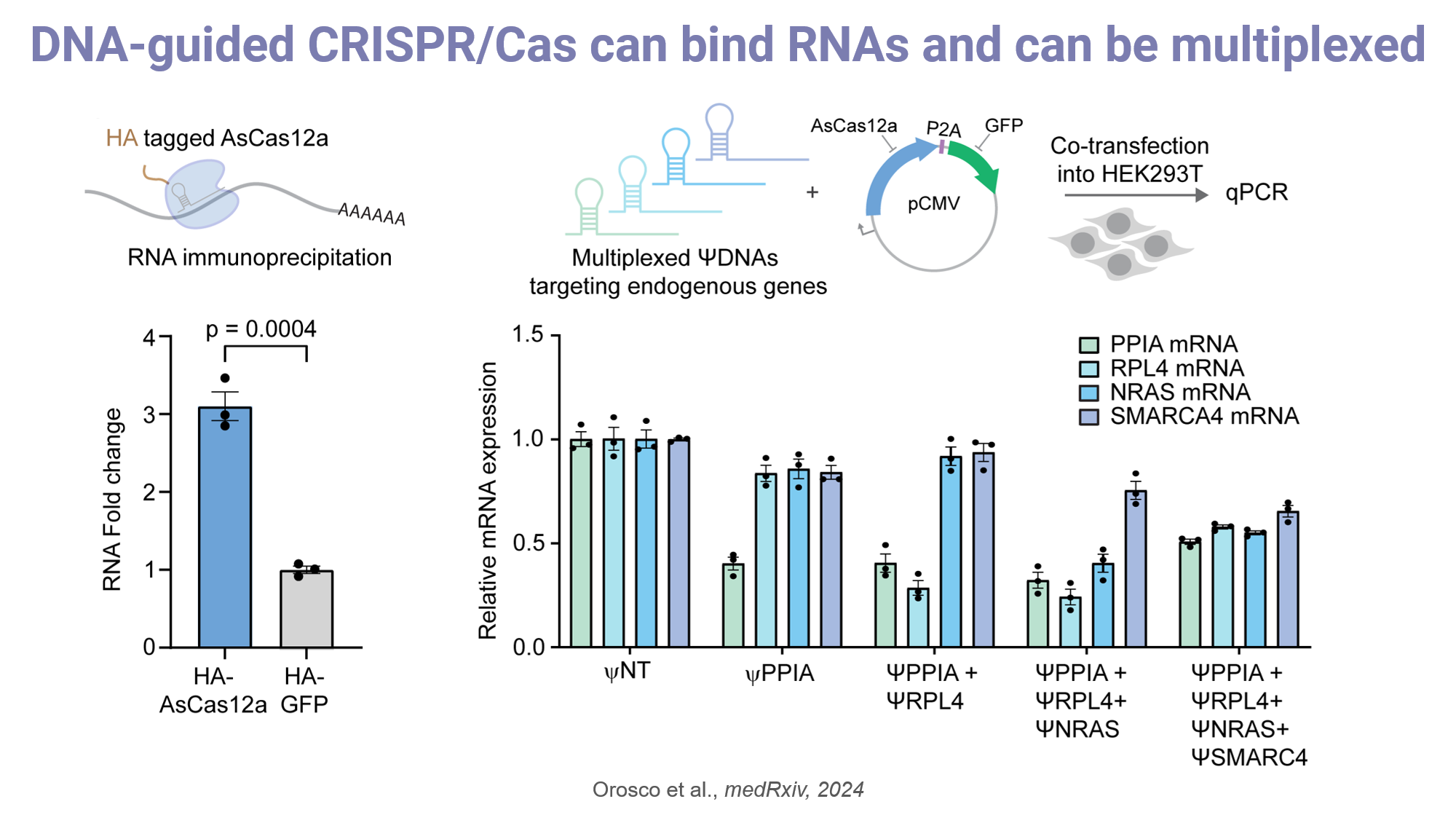
Next, in HEK293T cells, co-delivery of AsCas12a and ΨDNAs targeting mCherry mRNA resulted in significant translational repression through ribosome stalling, confirmed via microscopy, flow cytometry, and RT-qPCR. Modified ΨDNAs with phosphorothioate bonds or LNAs enhanced stability and hybridization, further increasing knockdown efficiency. To assess the system's broader applicability, we targeted endogenous genes (PPIA, RPL4, PCSK9, NRAS, SMARCA4). Remarkably, knockdown efficiencies robustly reached 95-99% for multiple targets in Cas12a-expressing HEK293 cells, outperforming many RNAi and Cas13 approaches [1]. Using combinations of ΨDNAs, we also demonstrated efficient multiplexed RNA knockdown across up to four targets simultaneously [3]. Furthermore, compared to Cas13, our system exhibited markedly lower off-target cleavage, as validated by RNA-Seq analysis [2]. This advantage positions DNA-guided Cas12a as a safer alternative for transcriptome engineering. Mechanistically, Cas12a lacks HEPN domains and does not cleave RNA directly; instead, ΨDNA guides mediate specific binding, which triggers downstream functions via attached effectors or fluorescence signal via trans-cleavage of DNA reporters. This binding-only approach ensures precision and minimizes unintended transcriptome disruption. To further enhance functionality, we fused AsCas12a with various effector domains enabling robust regulation RNA degradation, methylation, and base editing in cells. In an mCherry reporter system, ΨDNA-mediated base editing restored fluorescence, confirming programmable RNA editing [7].
In conclusion, ΨDNA-guided Cas12a represents a new RNA-targeting paradigm with a broad range of applications, from clinical diagnostics [8] to programmable gene regulation. It is cost-effective, highly modular, and suited for multiplexed gene control or therapeutic development. Its superior specificity, low toxicity, and flexible design make it a promising alternative to current RNA-targeting systems, extending the CRISPR toolkit for synthetic biology and biochemical engineering.
References:
- Abudayyeh, O. O. et al. (2017). RNA targeting with CRISPR–Cas13. Nature, 550(7675), 280–284. https://doi.org/10.1038/nature24049
- Apostolopoulos, A. et al. (2024). dCas13-mediated translational repression for accurate gene silencing in mammalian cells. Nature Communications, 15, 2205. https://doi.org/10.1038/s41467-024-40138-z
- Orosco C, Rananaware SR, Huang B, et al. (2024). DNA-guided CRISPR/Cas12 for RNA targeting. medRxiv, https://doi.org/10.1101/2024.11.21.24317744.
- Nguyen, L. T., Smith, B. M., & Jain, P. K. (2020). Enhancement of trans-cleavage activity of Cas12a with engineered crRNA. Nature Communications, 11, 4906. https://doi.org/10.1038/s41467-020-18770-8
- Rananaware, S. R. et al. (2023). Programmable RNA detection with CRISPR-Cas12a. Nature Communications, 14(1), 5409. https://doi.org/10.1038/s41467-023-41294-z
- Rozners, E. (2022). Chemical modifications of CRISPR RNAs to improve gene-editing activity and specificity. Journal of the American Chemical Society, 144(29), 12584–12594. https://doi.org/10.1021/jacs.2c04233
- Cox, D. B. T. et al. (2017). RNA editing with CRISPR-Cas13. Science, 358(6366), 1019–1027. https://doi.org/10.1126/science.aaq0180
- Chen, J. S. et al. (2018). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science, 360(6387), 436–439. https://doi.org/10.1126/science.aar6245