2025 AIChE Annual Meeting
(271e) Engineering Plant Immunity Proteins in Yeast: Rational Design to Overcome Limitations Imposed By Glycosylation
Authors
Changing protein function requires changing protein sequence. However, most changes made to proteins (called mutations) will worsen function. Thus, many mutations need to be tested to identify those that are beneficial and filter out those that are harmful. Mutagenesis studies performed in plants are time-consuming and low-throughput due to the long life cycles and low transformation efficiencies of plants. Yeast surface display (YSD) offers a promising alternative that allows for high-throughput screening of much larger libraries of mutations.
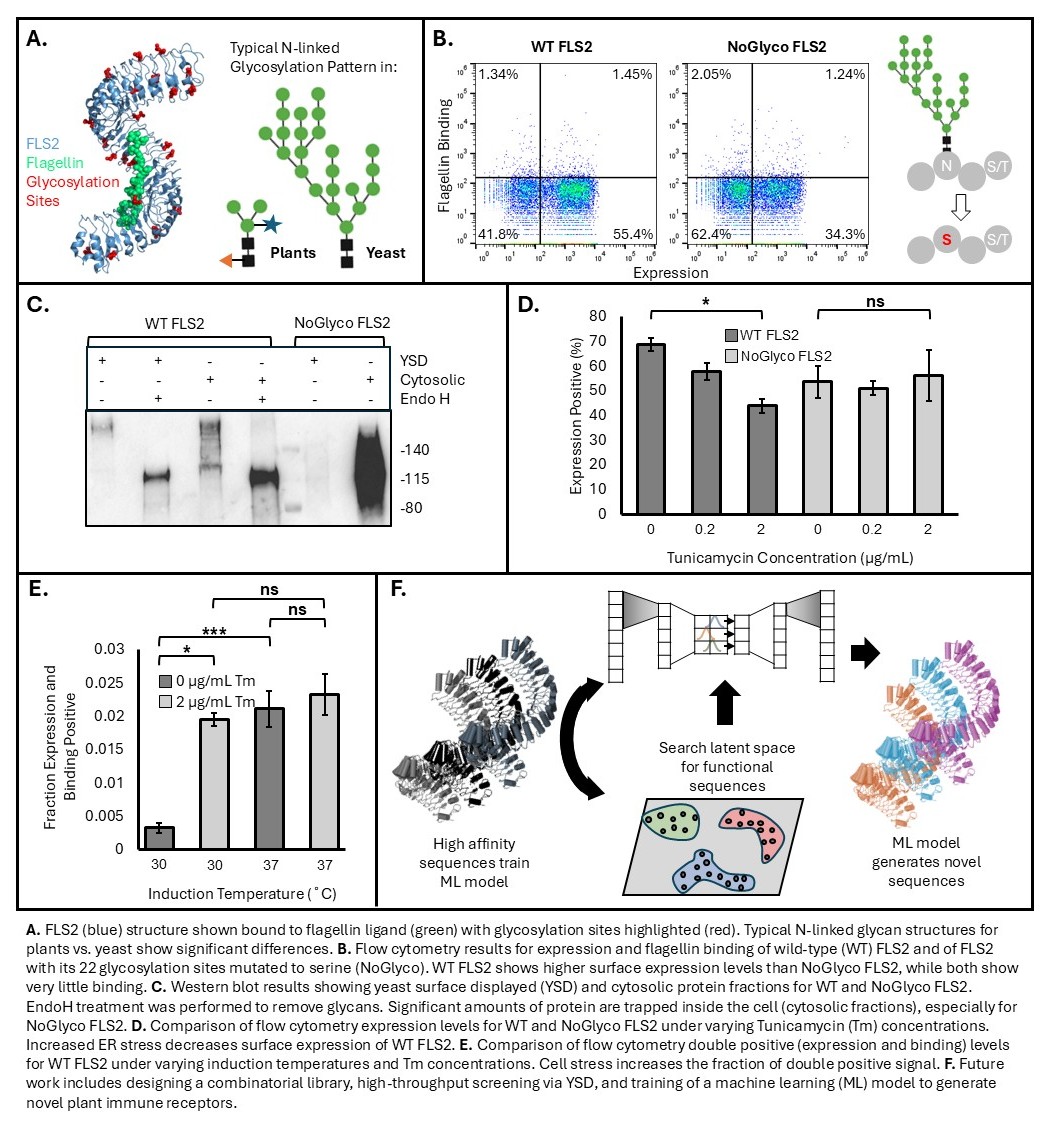
We have expressed FLS2 in S. cerevisiae, achieving >55% surface expression levels in flow cytometry assays. Further investigation into yeast surface displayed FLS2 shows that the protein is not functional, with <2% of the population binding flagellin. A key challenge in functional FLS2 expression are the differences between plant and yeast glycosylation, a post-translational modification in which sugars are attached to the protein which may affect structure and function. We have observed that very large sugars are added to the protein by the yeast, making it look very different from when it is made in plants. We are investigating the effect of these sugars on FLS2, including their impact on defense properties as well as limitations they may have on protein production.
To examine the effects of removing glycosylation from the protein, all 22 glycosylation sites were mutated from asparagine to serine, preventing sugar attachment (this mutated version is referred to as “NoGlyco FLS2”). Expression levels on the surface of yeast are lower (~35%) for NoGlyco compared to wild-type (WT) FLS2, potentially indicating the structural importance of glycosylation, with no significant change in binding. When treated with tunicamycin (Tm), a potent inhibitor of N-linked glycosylation, expression levels decrease for WT but not NoGlyco FLS2. This further supports the importance of glycosylation for WT FLS2 expression, while NoGlyco FLS2 – a non-glycoprotein – is not affected. Stressing the cell, either through Tm treatment to induce ER stress or increased temperature to induce thermal stress, resulted in a >5-fold increase in flagellin binding for WT FLS2. Under stress, only the most stable proteins are sent to the cell surface, so an increase in binding is observed, while overall expression decreases. Next, we plan to investigate FLS2 variants with subsets of glycosylation sites mutated to inform the degree of glycosylation necessary for stability and function. Additional investigation into FLS2 stability and structure will also be performed through differential scanning fluorimetry and circular dichroism spectroscopy.
Following functional FLS2 expression, future work will include designing a combinatorial library by mutating a panel of positions in and around the binding pocket. High-throughput screening of the yeast surface display library will be performed using magnetic bead sorting and fluorescence-activated cell sorting. These sequences will be used to train a machine learning model, such as a variational autoencoder, to generate new FLS2 sequences. If successful, this method will be expanded to other plant immunity proteins. The development of a platform to reliably engineer plant proteins will enable the generation of disease-resistant crops, improving food security.