2025 AIChE Annual Meeting
(288c) Award Submission: Sensitive Spatial miRNA Profiling of Plant Tissue Using Spectral Rolling Circle Amplification (RCA) Multiplexing Techniques in Nanoliter Well Arrays
Authors
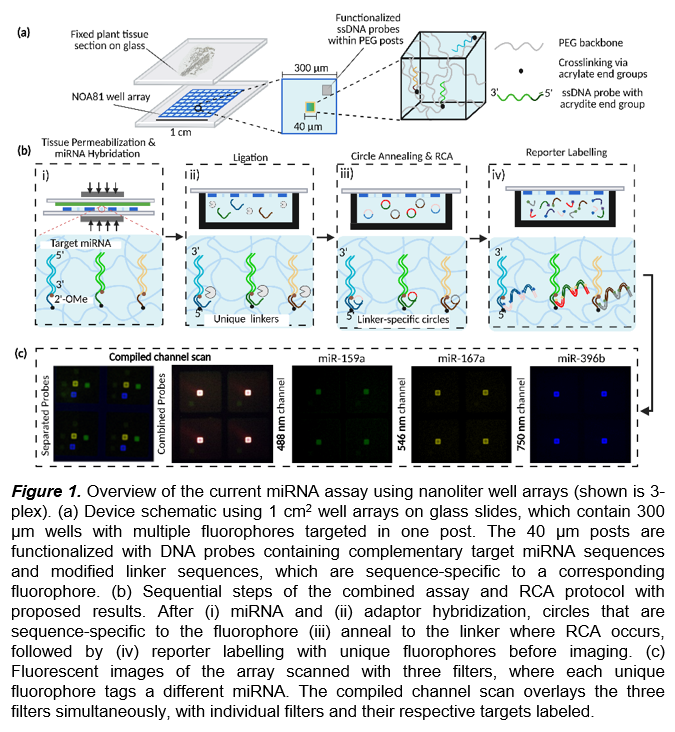
We use a nanoliter well array with current well feature sizes of 300 μm x 300 μm to isolate sections of the fixed plant tissue (10 μm thick) (Fig. 1a). Hydrogels crosslinked as square posts are evenly spaced within each well of the array and UV-photopatterned using contact lithography. We functionalize each post containing a miRNA-specific probe. These DNA probes consist of a complementary miRNA strand and a universal adapter sequence. After posts are fabricated, the array is sandwiched between a fixed plant tissue section and hermetically sealed using magnets. Reagents developed to free miRNA by simultaneously disrupting tissue and permeabilizing cells then hybridize to the complementary miRNA target. To quantify the amounts of miRNA in the well array, the current detection scheme utilizes R-Phycoerythrin (R-PE) fluorophores to determine the relative position of the hydrogel posts within each well, which corresponds to the target miRNA. We previously demonstrated that hydrogels embedded with the complementary miRNA probe sequence are utilized to achieve spatially resolved, 3-plexed miRNA detection, represented as a heatmap. These miRNA spatial patterns conform to the tissue geometry and can discriminate between different accumulation patterns, enabling region-specific quantification within Arabidopsis leaves.
Further, we will detail the expansion of our platform using spectral detection techniques with different fluorophores, along with rolling circle amplification (RCA), to enhance the existing sensitivity and multiplexing capability for miRNA capture (Fig. 1b). Using a single fluorophore (R-PE) to locate each miRNA spatially limits the number of unique targets that can be captured in the array by reducing the physical space in each well. To overcome this limitation, we demonstrate the integration of five unique fluorophores within each post in our assay workflow, motivated by the initial evaluation of fluorescence spectra to minimize overlap between fluorescent dyes. Each fluorophore is selected by its extinction coefficient and fluorescence quantum yield to determine its respective brightness. By using narrow-band filters to scan each fluorophore sequentially, we can incorporate unique, non-overlapping commercial dyes, such as Alexa Fluor and Atto (Fig. 1c). Therefore, simultaneously adding multiple colors to quantify the amount of miRNA present can increase the multiplexing by at least five times the current amount with minimal modification to the current assay.
In addition to increasing the number of detectable miRNA targets, having high sensitivity of nucleic acids is a necessary means to understand the fundamental roles of various biomolecules in low amounts. Signal amplification of nucleic acids enables greater sensitivity for detecting low-abundance transcripts. Rolling circle amplification (RCA) is a technique that isothermally amplifies DNA to generate a linear sequence for the addition of multiple fluorescent reporters to hybridize on an extended strand. Since plants have larger cells and contain less miRNA, RCA is pivotal in expanding the capabilities of target readout to discover the spatial position of less abundant miRNA. The Doyle group previously incorporated RCA in nanoliter well arrays, where RCA was applied to generate a 100-fold increase in sensitivity using SA-PE in the zeptomole detection regime, with a limit of detection (LLOD) of ~100 molecules. We also observe an increase in the number of fluorophores bound to each linker, resulting in a 50-fold improvement in the LLOD for each target.
Due to the flexibility of our assay, we will integrate RCA with spectral multiplexing by initially modifying the DNA probe sequence to target a unique fluorophore downstream. After miRNA hybridization, the unique adaptor, paired with detecting a specific fluorophore, is ligated to the RNA-DNA complex. Circular annealing using sequence-specific circular templates occurs where all circles are hybridized simultaneously. The circular template sequences are generated such that the circular annealing is specific to the adapter sequence and hybridization of reporters occurs with only fixed conjugation to different fluorophores. We fabricate DNA circles by using a linear template (50 bp) and CircLigase to conjugate the two ends of the linear template. Circle synthesis is then validated using spectrophotometry techniques (NanoDrop) and running gel electrophoresis to consider the products as a pure nucleic acid sample. After circular annealing, RCA will be initially implemented by expanding all circles, carried out under optimized conditions previously developed. Labelling after RCA occurs with reporters containing pre-conjugated fluorophores for direct imaging. After performing the RCA assay, we image using the same fluorescent scanner with predetermined parameters set according to the spectral characteristics of the fluorophores.
We will also examine the signal obtained with miRNA amounts previously undetectable without RCA and validate the assay by comparing the signal directly with the non-amplified miRNA amounts. Demonstration of the amplified spectral multiplexing technique will be applied to A. Thaliana leaves, where prior work in the Doyle group showed successful plant tissue permeabilization and spatial miRNA detection from four-week-old A. Thaliana leaves. Therefore, we will demonstrate that integrating the nanoliter well array platform by enhancing target amplification (RCA) and spectral multiplexing (utilizing five unique fluorophores) can increase the number of targets detected by an order of magnitude, with a LLOD of ~100 molecules, as shown using A. Thaliana leaves. Our results showcase a significant step towards increased sensitive multiplexing, as prior spectral amplification work has been leveraged with RCA using two colors (4-plex) in both solution and on DNA microarrays. We can expand our platform to incorporate higher sensitivity using RCA by directly capturing miRNA using surface-based probes. We demonstrate that integrating additional fluorophores for specific target labelling can increase multiplexing by at least 35-plex with five colors, showcasing minimal changes to the existing workflow while imparting high detection specificity.
Using our technique, we can combine different DNA probes to capture multiple miRNA types in one singular post for target differentiation with sequential fluorophore channel scanning. By integrating spectral multiplexing with RCA, we can increase resolution by decreasing well feature sizes towards single-cell resolution, offering a highly precise and sensitive toolkit for plant biologists to understand spatial changes across fixed plant tissues.