2024 AIChE Annual Meeting
(101c) AI Driven Immune Mapping of Breast Cancer
Authors
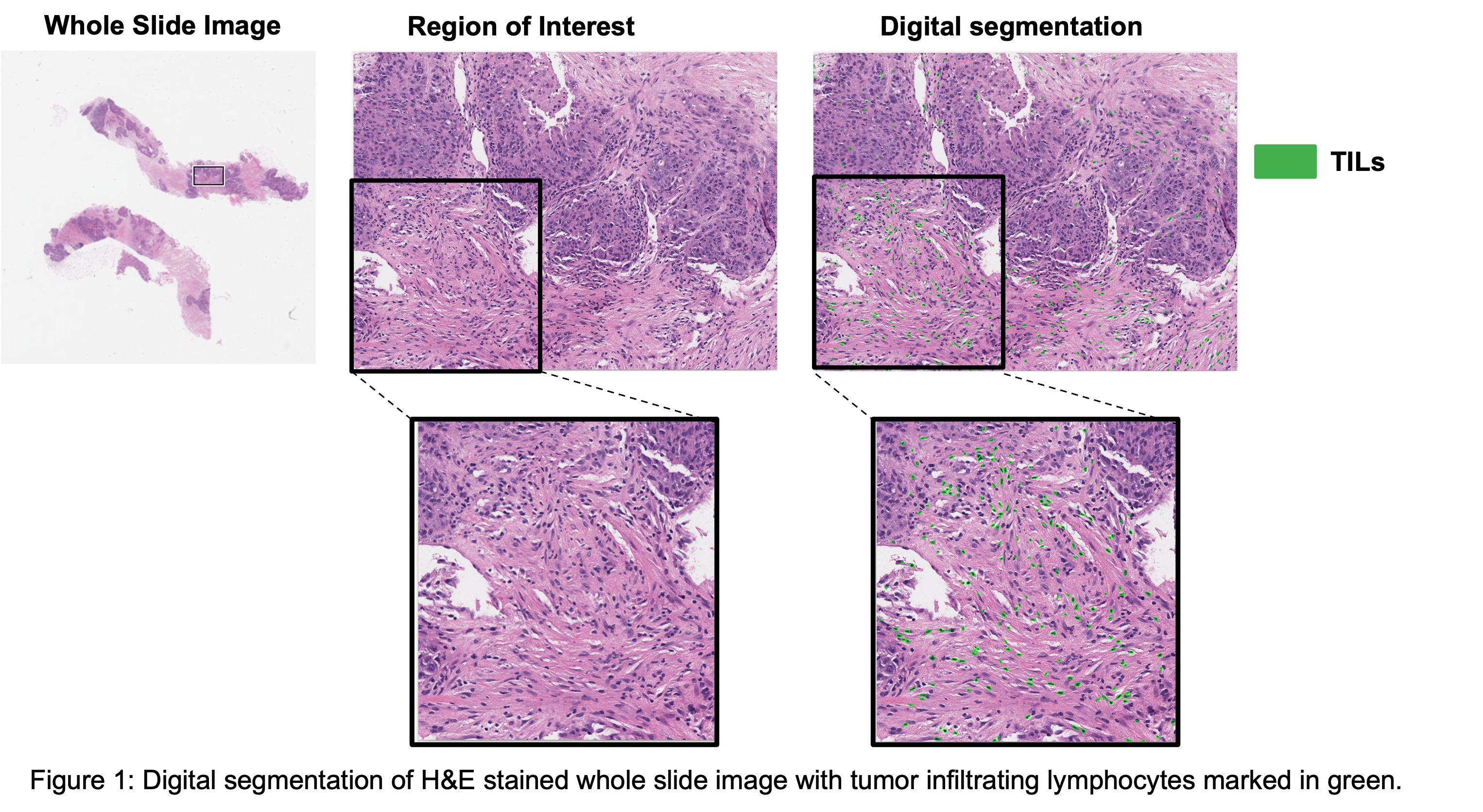
Methods and Results: We first identified a cohort of patients with locally advanced TNBC who underwent diagnostic biopsy and breast MRI prior to NAC. We have acquired both multiplex and pathology imaging data for the immune profiling of patients. We have utilized a combination of supervised and unsupervised machine learning models to digitally segment immune cells and other diagnostic regions of the biopsy. To overcome the limitation of access to labeled data, we have utilized clustering approaches to segment the data and combine it with simpler convolutional neural networks to minimize the user-based subjective interpretation of clustering results. Additionally, we have developed a sampling strategy that minimizes the impact of patient data variability on model generalizability. We have also compared and fine-tuned deep learning-based approaches for high-throughput cell segmentation of multiplexed imaging data. Concordance correlation coefficients were calculated between automated and pathologist measurements in addition to correlating the digital signatures with patient outcome data (response to treatment and recurrence). The resulting pipeline navigates through whole slide images, segments cells, and regions of interest in an automated, high throughput, and accurate manner. Our pipeline also generates a digital stromal TILs score, as shown in the figure below, which is then used as a prognostic marker for patient stratification.
Conclusions: This work is an interdisciplinary effort combining pathology, radiology, data science, and computer vision techniques. We have developed a standardized framework for TILs assessment that can assist pathologists in consistently using this metric for patient prognosis and treatment decision-making. Finally, we have combined the power of multimodal data (three types of imaging data, patient data from medical records) with AI to develop digital indicators of patient prognosis, particularly response to treatment and recurrence free survival.