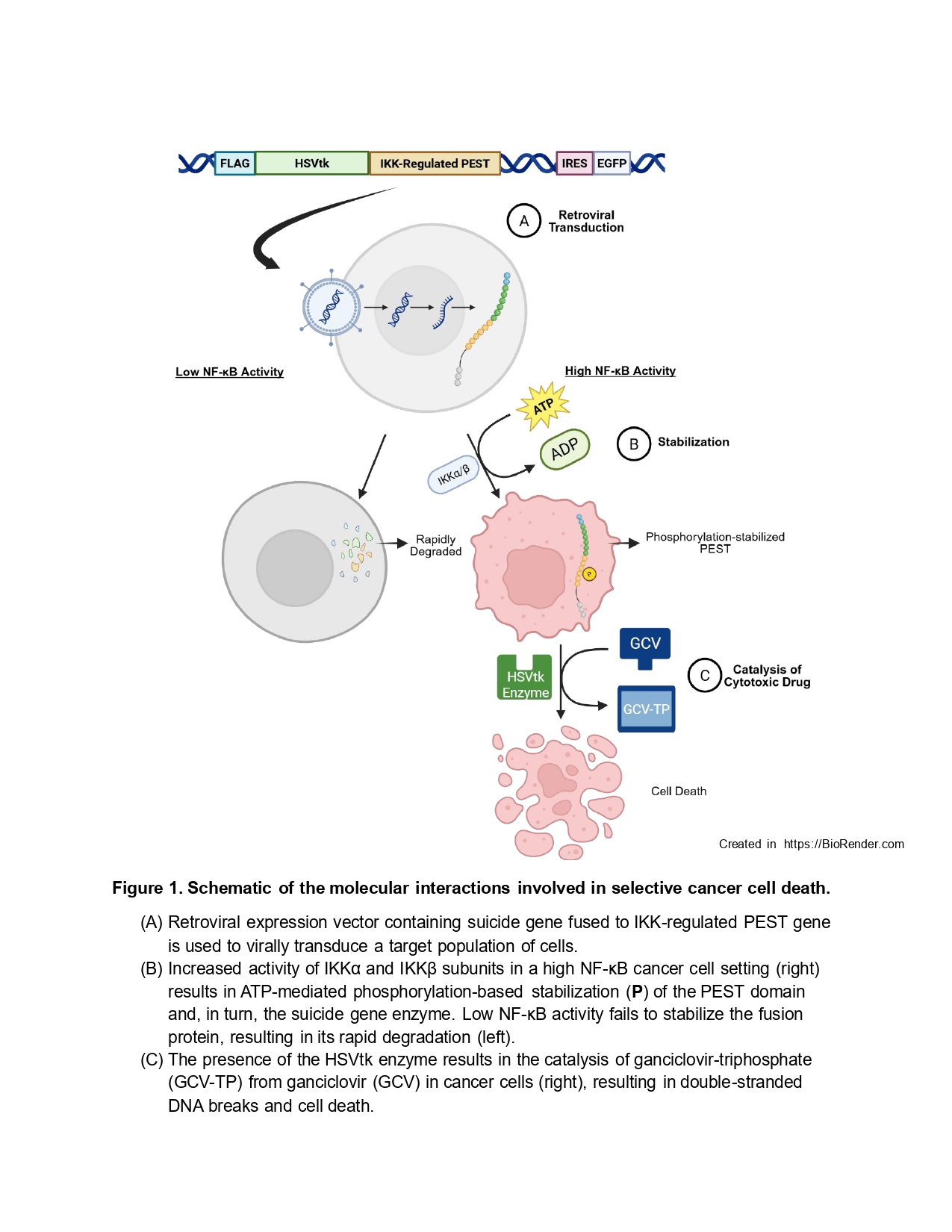
Suicide gene therapy (SGT) is a cutting-edge cancer treatment approach wherein virally encoded transgenes drive the cancer-cell expression of exogenous enzymes that convert innocuous prodrugs into cytotoxic products, thereby causing cell death. Selective and robust transgene expression in cancer cells has been a bottleneck in SGT, though, due to the insufficient activity of the cancer-specific promoters that have been tested to drive transgene expression.
1 Because of these limitations, no SGTs have yet been approved by the Food and Drug Administration (FDA). We previously improved on SGT designs by fusing the prodrug-converting enzyme herpes simplex virus thymidine kinase (HSVtk) with a PEST domain from the FRA-1 transcription factor, which is phosphorylated by the oncogenic extracellular signal-regulated kinase (ERK).
2 PEST domains are peptide sequences rich in proline, glutamic acid, serine, and threonine whose proteolytic turnover is typically greatly slowed by phosphorylation. Thus, our design created a novel HSVtk fusion that was driven by a strong viral promoter but selectively post-translationally stabilized in cancer cells where ERK was overactive.
2 This design is uniquely advantageous over alternative SGT designs because it capitalizes on the ability of cancer cells to rescue themselves from death via ERK activation in response to cytotoxic stress. While it would be useful to develop new versions of the kinase-stabilized SGT that leverage the activity of other cancer-relevant signaling pathways, surprisingly little is known about endogenous PEST domains that are specifically stabilized by kinases other than ERK. The objective of this research is to demonstrate that SGTs stabilized by other kinases can be developed and optimized by using data-driven modeling to guide protein engineering. Specifically, we sought to create a SGT stabilized by the activity of NF-κB (nuclear factor-κB) pathway. NF-κB pathway activity antagonizes apoptosis and promotes chemotherapeutic resistance in many cancer cell types. NF-κB is particularly relevant in pancreatic cancer,
3 which has a five-year survival rate of only ~13% and is characterized by unusual chemoresistance.
4 To build a SGT regulated by NF-κB, we focused on the inhibitor of κB kinases (IKK). The IKK subunits IKKα and IKKβ are activated by NF-κB pathway inducers and are frequently overactive in cancer.
5 Thus, IKK substrates are ideal candidates to study when trying to engineer PEST domains that will be stabilized in a cellular context of high NF-κB activity. Importantly, there are no reported IKK-regulated substrates with PEST functionality in the literature. As a result, we set out to engineer a small library of potentially IKK-regulated PEST domains and used a data-driven modeling approach to guide their initial cloning and testing. Twenty-five candidate domains were first designed by scoring five purported IKK substrates (insulin receptor
6, IκB
7, transcription factors IRF3 and IRF7
8, and CSN9
9)
for all possible phosphorylation sites and their affinities for IKKα- and IKKβ-mediated phosphorylation in the Kinase Library.
10 Peptide sequences surrounding residues targeted by phosphorylation were extracted, and sequence variations were generated for increased IKK specificity and codon-optimized for expression in human cells. A set of continuous features describing sequence, positional, and physiochemical information for first-generation regulatory protein designs was used to characterize the sequences along 12 dimensions. Principal component analysis (PCA) was then used to identify six first-generation regulatory proteins displaying good variance from one another within the 12-dimensional feature space. These six prospective IKK-regulated PEST domains were cloned into retroviral expression vectors as fusions with HSVtk. The selected fusions are being tested for stable expression and responsiveness to NF-κB pathway activation or inhibition in an HPAF-II pancreatic cancer cell background. Protein expression data from those tests will be used to generate a partial least squares regression (PLSR) model, which will identify the most important peptide features to tune for the creation of second-generation fusion designs with improved stability and IKK-dependent regulation. Successful implementation of this pipeline is anticipated to produce an optimized second-generation suicide gene construct, which will be tested in vitro and eventually in vivo in preclinical models of pancreas cancer.
1. Karjoo, Z.; Chen, X.; Hatefi, A. Progress and Problems with the Use of Suicide Genes for Targeted Cancer Therapy. Advanced drug delivery reviews 2016, 99 (Pt A), 113–128. https://doi.org/10.1016/j.addr.2015.05.009.
2. Day, E. K.; Campbell, A.; Pandolf, A.; Rogerson, T.; Zhong, Q.; Xiao, A.; Purow, B.; Lazzara, M. J. ERK-Dependent Suicide Gene Therapy for Selective Targeting of RTK/RAS-Driven Cancers. Molecular Therapy 2021, 29 (4), 1585–1601. https://doi.org/10.1016/j.ymthe.2020.12.019.
3. Li, Q.; Yang, G.; Feng, M.; Zheng, S.; Cao, Z.; Qiu, J.; You, L.; Zheng, L.; Hu, Y.; Zhang, T.; Zhao, Y. NF-ΚB in Pancreatic Cancer: Its Key Role in Chemoresistance. Cancer Letters 2018, 421, 127–134. https://doi.org/10.1016/j.canlet.2018.02.011.
4. Hegazi, A.; Rager, L. E.; Watkins, D. E.; Su, K.-H. Advancing Immunotherapy in Pancreatic Cancer. International Journal of Molecular Sciences 2024, 25 (21), 11560. https://doi.org/10.3390/ijms252111560.
5. Mercurio, F.; Manning, A. R. NF-ΚB as a Primary Regulator of the Stress Response. Oncogene 1999, 18 (45), 6163–6171. https://doi.org/10.1038/sj.onc.1203174.
6. Gao, Z.; Hwang, D.; Bataille, F.; Lefevre, M.; York, D.; Quon, M. J.; Ye, J. Serine Phosphorylation of Insulin Receptor Substrate 1 by Inhibitor ΚB Kinase Complex. Journal of Biological Chemistry 2002, 277 (50), 48115–48121. https://doi.org/10.1074/jbc.m209459200.
7. Israel, A. The IKK Complex, a Central Regulator of NF- B Activation. Cold Spring Harbor Perspectives in Biology 2009, 2 (3), a000158–a000158. https://doi.org/10.1101/cshperspect.a000158.
8. Antonia, R. J.; Hagan, R. S.; Baldwin, A. S. Expanding the View of IKK: New Substrates and New Biology. Trends in Cell Biology 2021, 31 (3), 166–178. https://doi.org/10.1016/j.tcb.2020.12.003.
9. Zhang, J.; Zhao, R.; Bryant, C. L. N.; Wu, K.; Liu, Z.; Ding, Y.; Zhao, Y.; Xue, B.; Pan, Z.-Q.; Li, C.; Huang, L.; Fang, L. IKK-Mediated Regulation of the COP9 Signalosome via Phosphorylation of CSN5. Journal of proteome research 2020, 19 (3), 1119–1130. https://doi.org/10.1021/acs.jproteome.9b00626.
10. Johnson, J. L.; Yaron, T. M.; Huntsman, E. M.; Kerelsky, A.; Song, J.; Regev, A.; Lin, T.-Y.; Liberatore, K.; Cizin, D. M.; Cohen, B. M.; Vasan, N.; Ma, Y.; Krismer, K.; Robles, J. T.; van de Kooij, B.; van Vlimmeren, A. E.; Andrée-Busch, N.; Käufer, N. F.; Dorovkov, M. V.; Ryazanov, A. G. An Atlas of Substrate Specificities for the Human Serine/Threonine Kinome. Nature 2023, 1–8. https://doi.org/10.1038/s41586-022-05575-3.