2025 AIChE Annual Meeting
(389bs) Targeting Low-Energy Backmapped Protein Ensembles with Adjoint Matching
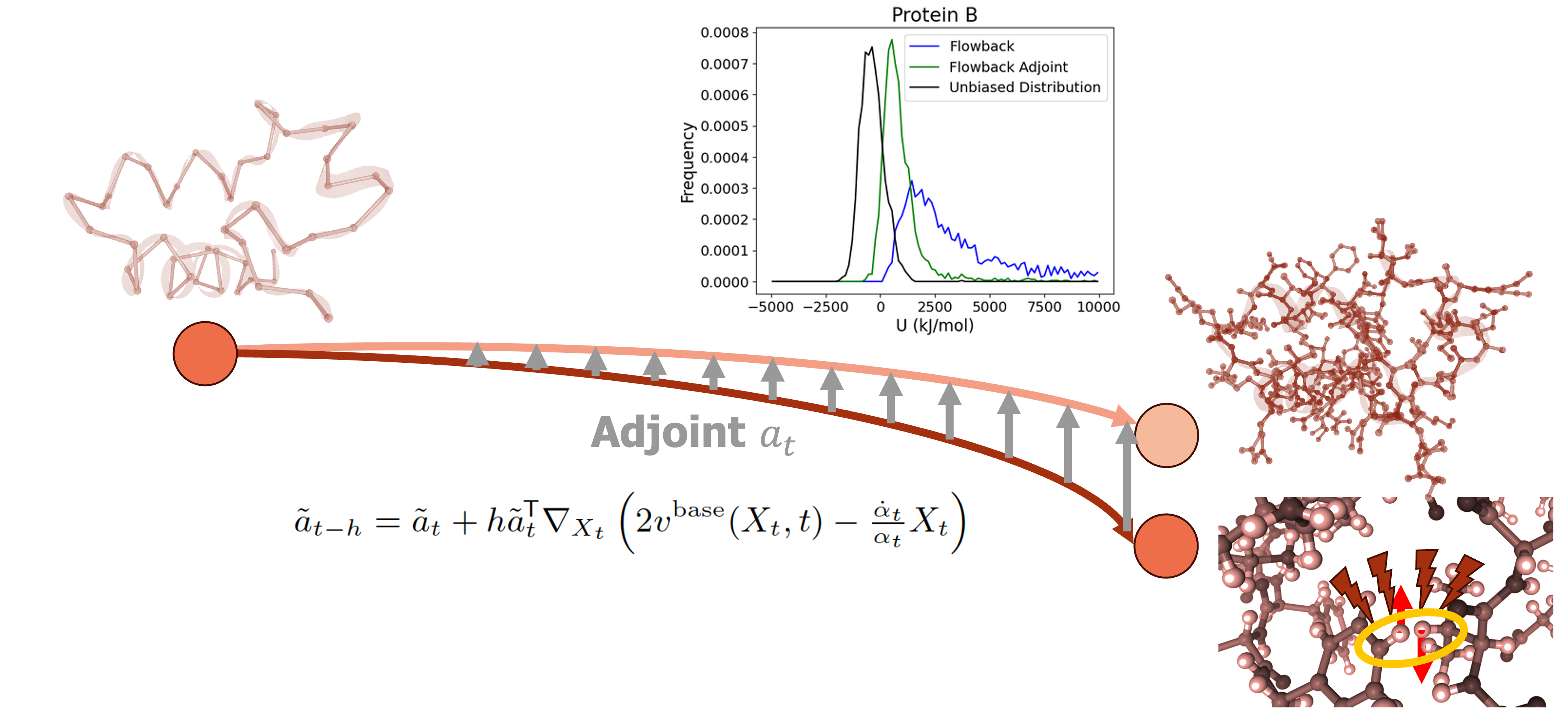
Backmapping reconstructs all-atom detail from coarse-grained simulations, requiring physically plausible structures and energetically favorable conformations that can be used to initialize molecular dynamics (MD) simulations. A robust backmapping approach should generate diverse, low-energy structures that reflect the ensemble of corresponding all-atom configurations. We build on the pretrained FlowBack model, using it as a prior and introducing an adjoint matching approach to improve the energetic quality of backmapped structures. Specifically, we post-train FlowBack with an energy-based reward that penalizes high-energy conformations, encouraging the model to generate structures that not only minimize steric clashes but also correspond to lower-energy minima under a molecular force field using the adjoint-matching framework to adjust the pretrained flow model at every timestep. We validate our method on near-millisecond DESRES trajectories, demonstrating that adjoint matching reduces the energy gap between backmapped and ground-truth structures while maintaining structural diversity. Our results suggest that integrating energy-based corrections into backmapping pipelines can improve the reliability of generated structures for MD simulations, offering a generalizable framework for enhancing coarse-grained-to-all-atom reconstruction.