2025 AIChE Annual Meeting
(190a) Structural Complexity Quantification of Network Materials Using a Graph Theoretical Approach
Authors
Kody Whisnant - Presenter, Wayne State University
Dickson Owuor, University of Michigan
Connor McGlothin, University of Michigan
Zi-Meng Han, University of Science and Technology of China
Mingqiang Wang, State Key Laboratory of Urban Water Resource and Environment Harbin Institute of Technology
Vinicius Alevato, Wayne State University
Stephanie L. Brock, Wayne State University
Yudong Huang, Harbin Institute of Technology
Shu-Hong Yu, University of Science and Technology of China
Nicholas A. Kotov, University of Michigan
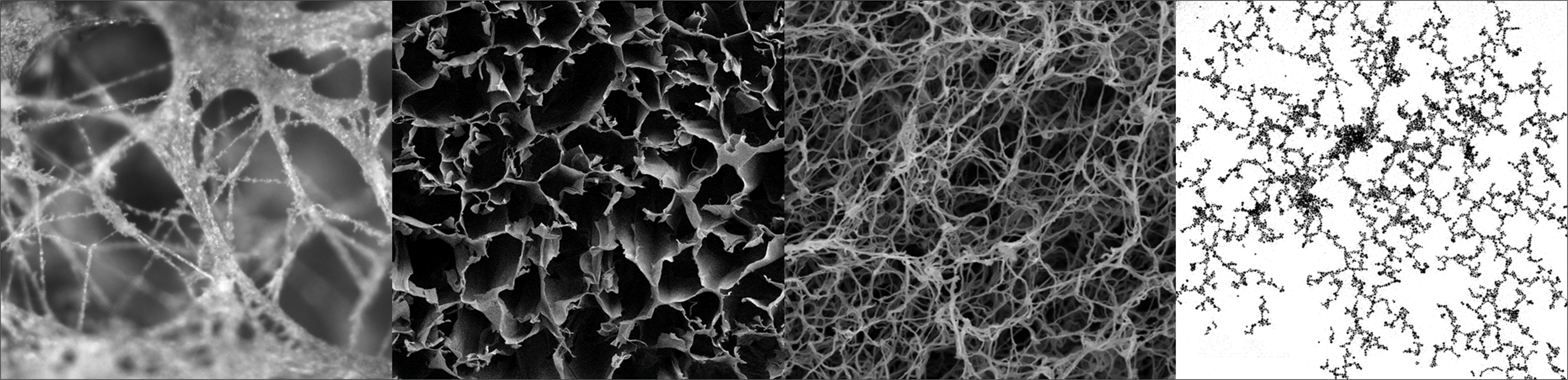
Networks, broadly encompassing systems of interconnected parts that exhibit structural properties, are all around us and include social relationships, the internet, highways, electrical power grids, subways, flight paths, neural connections, and chemical reactions. Materials with network-like morphologies are of notable importance in the fields of engineering, medicine, chemistry, and biology where their interconnected structure serves crucial functions. Structural complexity, or material architecture reflecting the combination of order and disorder, of networks spanning biological and inorganic materials is a crucial parameter controlling vital mechanical, thermal, electric, optical, chemical, and biological properties that determine material behavior. There is however no unified approach to measure complexity as prior studies largely inspired by biological systems were not adapted for the nanoscale structures currently being investigated. Graph Theory (GT) employs a systematic protocol to describe material networks through graph, G(n,e), models consisting of data points (nodes, n) joined by lines (edges, e). These graph representations allow for quantified characterization of material complexity through calculation of network theory descriptors of short- and long-range order that relate complexity and functionality, enabling the prediction of complex material properties required for effective materials design. This work utilizes a method of GT-based descriptor calculation to quantify structural complexity differences among network materials exhibiting a range of architectures.
Figure: Examples of structurally complex organic and inorganic networks.