2025 AIChE Annual Meeting
(383ag) Stochastic Simulation and Machine Learning Techniques for Studying Cell Differentiation and Maturation
My research focuses on applying the process system engineering principles to the biomanufacturing of cardiomyocytes, the contractile heart cells. Cardiomyocytes derived from human induced pluripotent stem cells (hiPSCs) can be used for drug testing, disease modeling, and regeneration therapies for cardiovascular diseases. However, two major challenges hinder their clinical and industrial adoption. (1) The structural immaturity of the hiPSC-derived cardiomyocytes, and (2) the Inability to consistently produce specific heart cell subtypes. My research addresses these challenges through the following approaches: (1) developing statistical and machine-learning-based methods to assess the structural maturity of differentiated cardiomyocytes, and (2) modeling cellular differentiation as a dynamic process to understand how cells respond to differentiation protocols.
In collaboration with experimental researchers, we investigated how the initial geometry of photo-cross-linkable polymer, called microspheroids (a mixture of PEG-Fibrinogen and hiPSCs), affect the structural maturity of the hiPSC-derived cardiomyocytes. We hypothesized that three parameters, the radial diameter, axial ratio (ratio of axial diameter to radial diameter) and the hiPSC concentration could affect the maturity-related features. High-resolution images of stained cardiomyocytes were captured, and structural features were extracted using Sarcomere Texture analysis and SarcTrack MATLAB tools. To assess cell maturity, I introduced two methods to identify potential mature cardiomyocytes. The first approach ranks cells based on individual maturity-related features and averages these ranks to compute a Structural Maturity Index (SM-Index). The second approach uses fuzzy clustering to group cells into two categories. Correlation analysis (Pearson and Spearman) indicated that the axial ratio showed a weak relationship with maturity features.
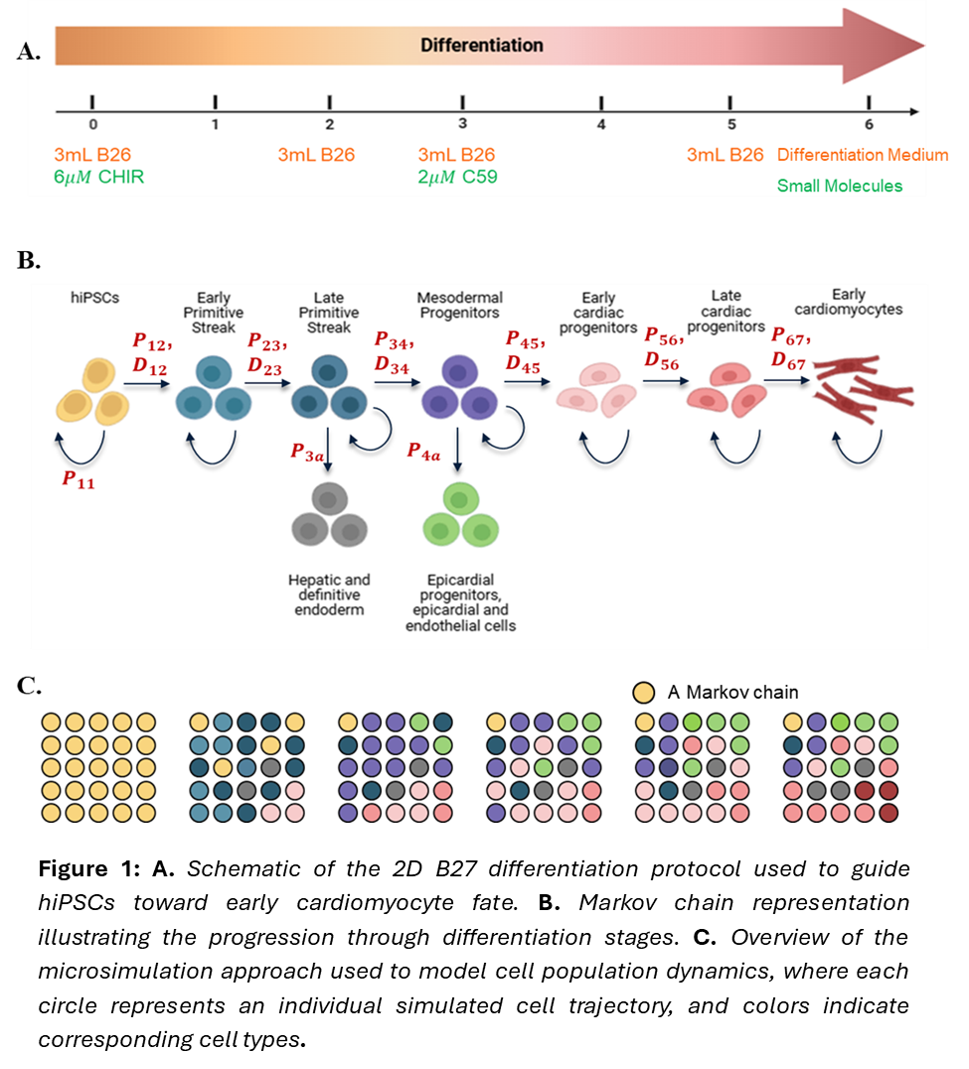
In my second project, I developed a simulation model to predict the cell type population changes during the early days of cardiac differentiation for a given protocol. I used a Markov chain and a microsimulation model to simulate the cell type population changes. The Markov chain simulates the differentiation of a single cell. The states in the Markov chain correspond to the cell types that are observed during the differentiation. The transition between the states is governed by the transition probabilities. The time spent by the cell before transitioning is sampled from an exponential distribution. To simulate population-level outcomes, the model runs 5,000 independent trajectories, recording each cell's state over time. These trajectories are aggregated to estimate cell population fractions on each day. I used Bayesian optimization to estimate both the transition probabilities and time distribution parameters. While the model captured key trends in cell population dynamics, the predictive performance ( =) was limited due to variability in the datasets used for parameter estimation. Future work will focus on improving robustness by accounting for cell line-specific variability and biological noise in the experimental data.
Across these projects, I have applied a wide range of process systems engineering tools, including microsimulation, Markov modeling, Bayesian optimization, machine learning, and image analysis, to solve problems in biological domain. My work demonstrates an ability to develop scalable, data-driven solutions, whether it's modeling stem cell differentiation, quantifying structural maturity in cardiomyocytes, or building image processing pipelines. These interdisciplinary experiences have strengthened my skills in algorithm development, data interpretation, and collaborative problem-solving, and I am excited to apply them to real-world challenges in research or industry.