2025 AIChE Annual Meeting
(369e) Short Peptides As Mixed-Mode Chromatographic Ligands: Establishing Design Rules
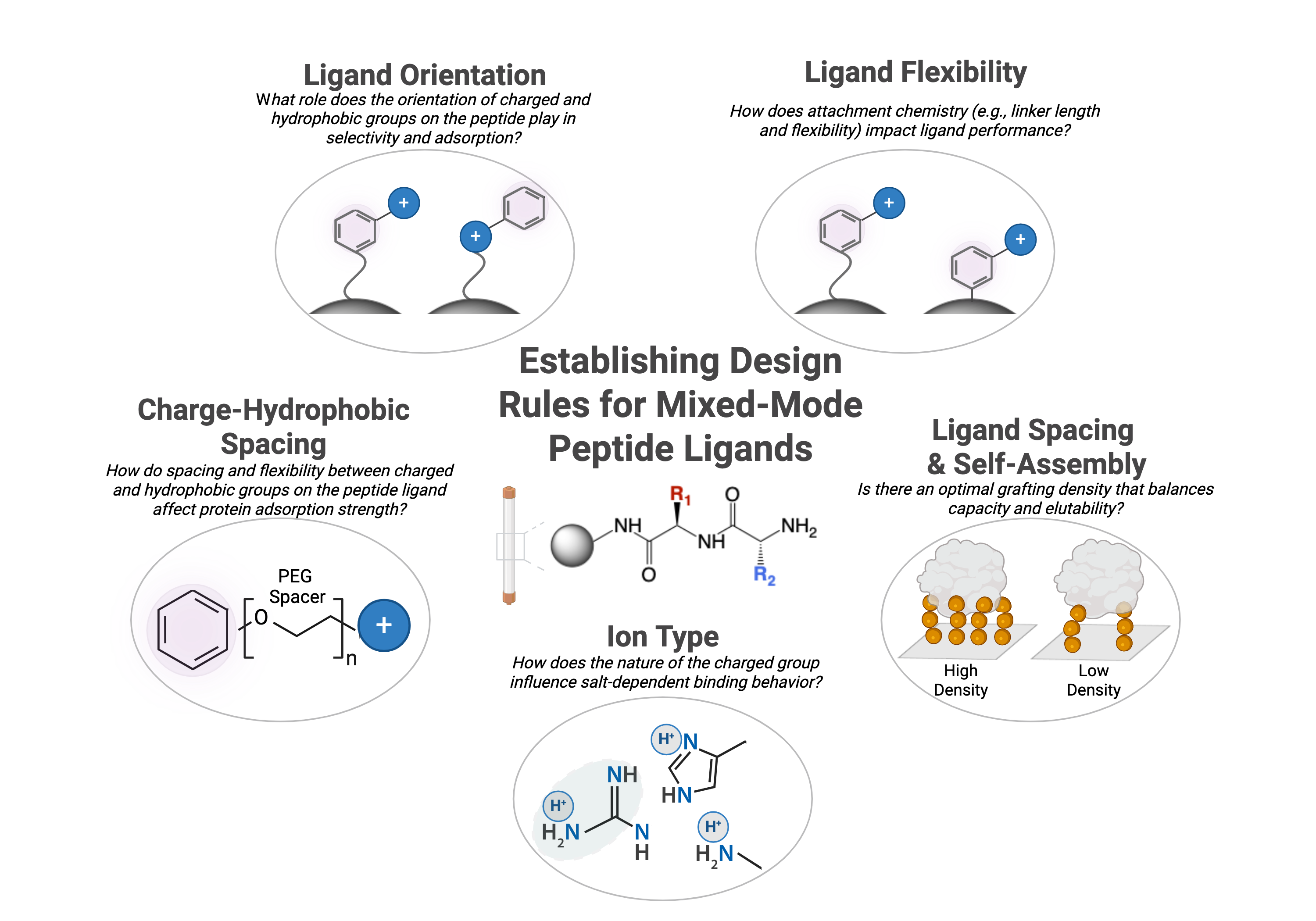
Peptides offer a promising platform for mixed-mode ligand design due to their chemical diversity and ease of synthesis directly on chromatographic resins. In this work, we synthesized homologous libraries of peptide-functionalized mixed-mode anion exchange resins to investigate several key design questions:
- How do spacing and flexibility between charged and hydrophobic groups on the peptide ligand affect protein adsorption strength?
- How does the nature of the charged group (e.g., lysine’s primary amine vs. arginine’s guanidinium) influence salt-dependent binding behavior?
- What role does the orientation of charged and hydrophobic groups on the peptide play in selectivity and adsorption?
- How does attachment chemistry (e.g., linker length and flexibility) impact ligand performance?
- Is there an optimal grafting density that balances capacity and elutability?
We synthesized and evaluated peptide ligand libraries against a panel of seven model proteins with diverse physicochemical properties. Results revealed that intermediate spacing between functional groups maximized adsorption strength—likely due to an interplay between spatial complementarity and conformational entropy. Interestingly, linker modifications affected ligand-resin performance differently, and to a much lesser extent, than ligand internal spacing. Peptide orientation (i.e., sequence directionality) had a marked effect on binding strength, and the identity of the charged group significantly influenced salt sensitivity. Finally, grafting density studies showed that while optimal densities were relatively consistent across ligands, molecular dynamics simulations suggest that ligand-ligand interactions and clustering may modulate protein binding behaviors. Together, these findings enhance our understanding of mixed-mode ligand design and lay the groundwork for next-generation chromatographic resins for emerging classes of biologics.