2025 AIChE Annual Meeting
(81b) Sequence-Dependent Study of Pentapeptide Self-Assembly with Molecular Dynamics
Methods: All-atom molecular dynamics (MD) simulations were performed using AMBER-compatible force fields (FF19SB) and the OPC water model with necessary salt ions to replicate physiological solvation. Eight peptide sequences were simulated, each with 30 copies in explicit solvent for at least 400 ns. Analyses were conducted using MDAnalysis in Python to quantify structural features such as clustering, π–π interactions (TYR–PHE, TYR–TYR, PHE–PHE), relative solvent-accessible surface area (relSASA), and C-terminal (tail-to-tail) distances. Contact matrices were built using a 4 Å heavy atom cutoff. Data were block-averaged and statistically analyzed over the final 200 ns of each simulation. To evaluate sequence-dependent assembly behavior, data were analyzed statistically across the last 200 ns.
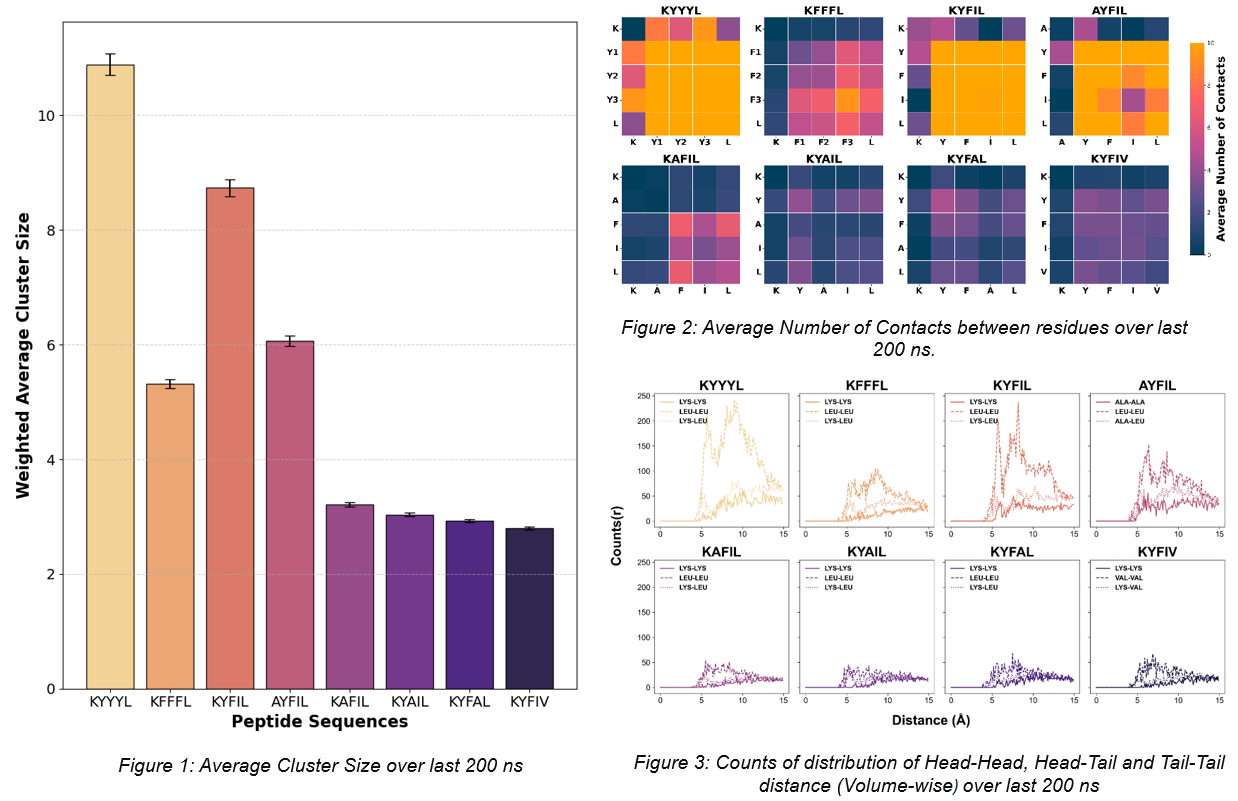
Results: Molecular dynamics simulations revealed distinct sequence-dependent differences in assembly behavior. A previous study has shown that KYFIL and AYFIL form hydrogels under physiological pH, whereas KYFIV, KYAIL, KYFAL, and KAFIL do not2. In our experimental study, KFFFL was observed to form a hydrogel. The gelation behavior of KYYYL has not yet been tested. Cluster size distribution analysis showed that peptide sequences KYFIL, KYYYL, AYFIL & KFFFL preferentially formed fewer, larger clusters, indicating strong assembly and potential for robust hydrogel network formation (Figure 1). In contrast, sequences like KYFIV, KYAIL, KYFAL and KAFIL formed smaller, more transient clusters, suggesting weaker self-association (Figure 1). Contact analysis between residues revealed that TYR–PHE , PHE-PHE and TYR–TYR interactions were enriched in sequences with strong gelation tendencies, suggesting the potential role of aromatic residues in stabilizing supramolecular structures (Figure 2). Although even 3000 ns simulations did not capture full nanostructure formation, analysis of tail-to-tail distance distributions showed distinct peaks in gelling peptides, suggesting early signatures of ordered packing relevant to hydrogel formation (Figure 3). If KYYYL is found to form a hydrogel in our experimental study, it would further strengthen our conclusions by providing a concrete link between experimental observations and computational predictions.
Conclusion: This work demonstrates the utility of atomistic MD simulations in revealing critical sequence–structure–function relationships in self-assembling pentapeptides. Key differentiating factors of gelling peptides include enriched aromatic interactions, larger cluster sizes, and characteristic tail-to-tail packing patterns. While atomistic MD is limited in fully capturing nanostructure (i.e. fiber, micelle) assembly as it is computationally intractable to perform long enough simulations to reach equilibrium, these findings do offer directional and mechanistic insight. Further exploration using coarse-grained models and generative diffusion-based design tools can accelerate the establishment of design for guiding novel ECM-mimetic peptide-based hydrogel discovery.
Acknowledgement: NSF DMR 2104723, UVA Research Computing
References:
1. Tang, J. D., Roloson, E. B., Amelung, C. D., & Lampe, K. J. (2019). Rapidly assembling pentapeptides for injectable delivery (RAPID) hydrogels as cytoprotective cell carriers. ACS Biomaterials Science & Engineering, 5(5), 2117–2121. DOI: 10.10 21/acsbiomaterials.9b00389
2. Tang, J. D.; Mura, C.; Lampe, K. J. A Stimuli-Responsive, Pentapeptide, Nanofiber Hydrogel for Tissue Engine J. Am. Chem. Soc. 2019, 141, 4886–4899. DOI: 10.10 21/jacs.8b13363
Keywords: Biomaterials, Peptide, Molecular Dynamics, Self-assembly