2025 AIChE Annual Meeting
(42b) Reducing Model Size and Improving Model Identifiability Using Model Based Design of Experiments
Authors
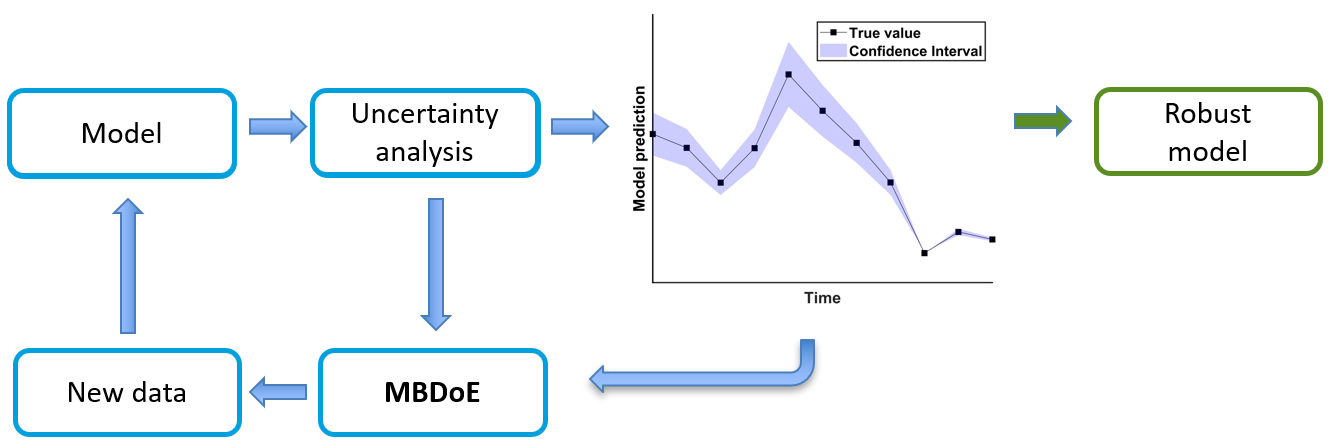
Model-Based Design of Experiments (MBDoE) offers a systematic framework for maximizing information gain while minimizing experimental effort. Unlike the traditional design of experiments, MBDoE leverages model structure and uncertainty to prioritize the most informative experiments. The utility of MBDoE requires expertise in statistics, computational optimization, bioprocess understanding and, modeling.
In this work, we applied identifiability analysis, a key component of MBDoE, to evaluate the adequacy of experimental data for parameter estimation of a dynamic metabolic flux analysis (DMFA) model of CHO cell metabolism for monoclonal antibody production. The initial DMFA model contained more than 50 kinetic parameters, many of which were difficult to estimate with the available data. Sensitivity analysis and the Fisher Information Matrix (FIM) were used to determine these unidentifiable parameters. The unidentifiable parameters were either fixed to reasonable values based on the model structure or eliminated to simplify the model.
The simplified DMFA model retained similar predictive capability while requiring fewer training data. The model was trained and validated using pseudo-perfusion and perfusion systems data, effectively capturing CHO cell metabolism across scales and operational modes.
By focusing on parameters with high uncertainty, subsequent experiments can be strategically designed using FIM based optimality criteria. These criteria - such as D-optimality or A-optimality—guide the selection of experimental conditions that help maximize the information gained about the uncertain parameters. The new experimental data combined with the existing one updates the model to re-evaluate parameter uncertainty. This iterative process- designing next experiments, updating the model, and reassessing uncertainty can improve model identifiability and robustness while reducing the required experiments. The outcome is a simplified yet predictive model with fewer, well-estimated parameters. This generalizable MBDoE framework enables efficient experimental planning to lower the cost and time associated with process development.