2025 AIChE Annual Meeting
(607b) A Predictive and Interpretable ML Framework to Guide Lipid Nanoparticle Design for Nucleic Acid Delivery
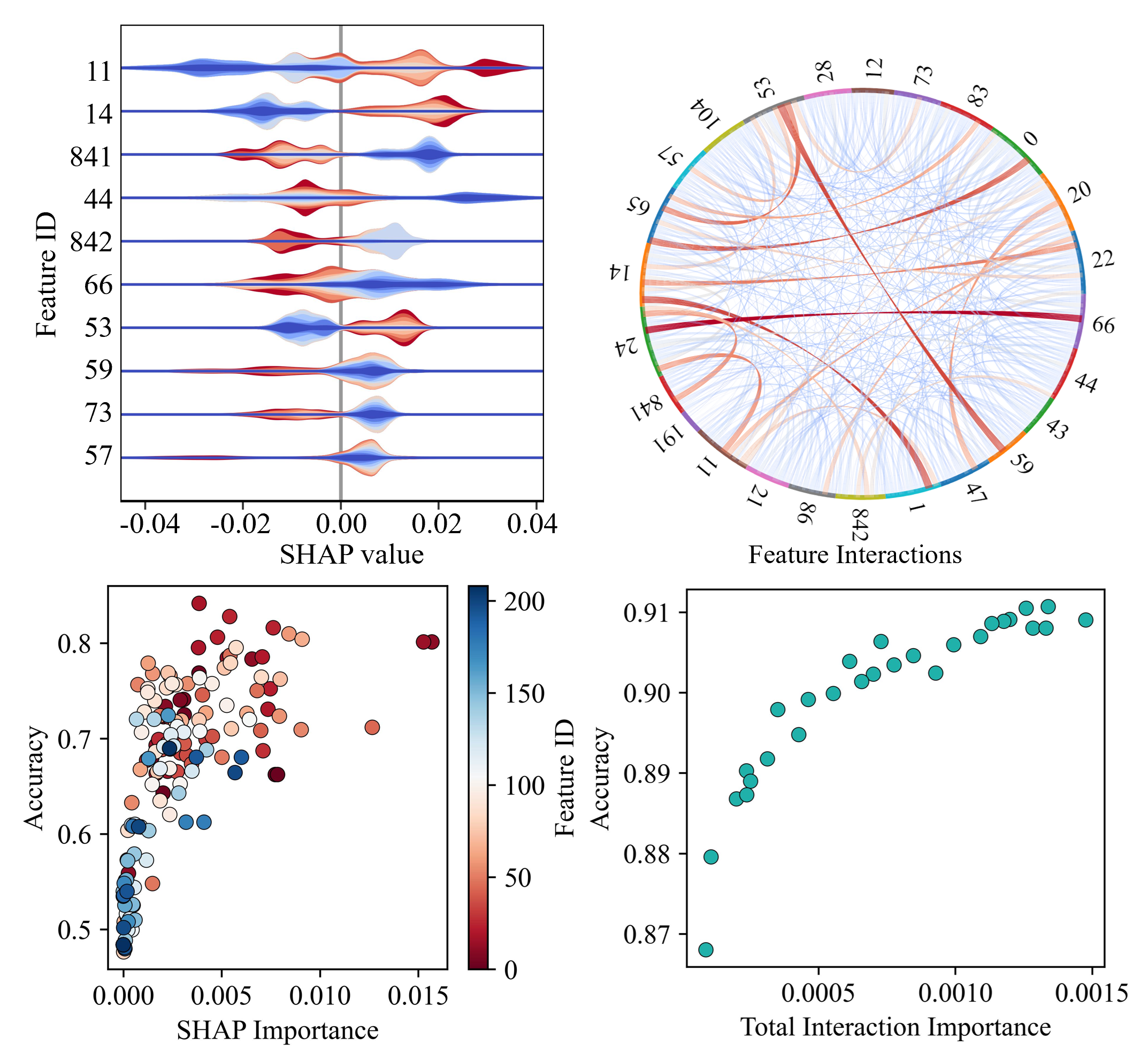
Lipid nanoparticles (LNPs) have emerged as highly effective carriers for gene therapies, including mRNA and siRNA delivery, owing to their ability to transport nucleic acids across biological membranes, low cytotoxicity, favorable pharmacokinetics, and scalability. A common approach to optimizing LNP design involves establishing quantitative structure-activity relationships (QSAR) between LNP composition and biological performance. However, developing such models is challenging due to the complexity of multi-component formulations, interactions with biological systems, and diverse physicochemical characteristics. To address these challenges, we developed a machine learning (ML) framework to predict the activity and cell viability of LNPs based on molecular and formulation features. Our curated dataset comprises 6,454 LNP formulations extracted from 21 independent studies. We implemented several molecular featurization techniques, ranging from physicochemical descriptors and structural fingerprints to graph-based representations, alongside six machine learning algorithms for both binary and multiclass classification tasks. Using scaffold-based five-fold cross-validation, the models achieved classification accuracies exceeding 90% for both activity and cell viability predictions. Among all model-feature combinations, descriptor-based features coupled with ensemble models such as balanced random forest and extra trees delivered the highest performance. A comprehensive SHAP-based feature analysis revealed that electrostatic descriptors, particularly those quantifying partial atomic charges on the ionizable lipid, were among the most influential features. Formulation-level parameters such as lipid-to-RNA ratio and dosage also emerged as critical predictors, underscoring the importance of composition and dosing strategy. Additional important features included descriptors capturing molecular shape complexity, topological indices, surface charge distribution, and lipophilic/hydrophilic surface area. Notably, our analysis showed that model performance was not solely governed by individual high-ranking features, but rather by synergistic interactions among complementary features. Clustering features based on SHAP interaction strengths revealed that groups of diverse physicochemical descriptors, including electrostatics, shape, lipophilicity, and formulation design parameters, collectively contributed more predictive power than individual features in isolation. Furthermore, we implemented a transfer learning strategy to incorporate in vivo-relevant features such as particle size, polydispersity index (PDI), and zeta potential into the predictive framework. Despite the smaller and imbalanced nature of the in vivo dataset, transfer learning models achieved accuracies exceeding 82%, demonstrating improved generalizability. This work highlights the potential of predictive and interpretable ML frameworks to guide rational LNP design and accelerate the development of nucleic acid delivery systems.