2025 AIChE Annual Meeting
(190f) Predicting Complexes between Chiral Nanoparticles and Proteins
Authors
Yanan Wang, Tianjin university
Emine Sumeyra Turali-Emre, University of Michigan
Neel Moudgal, University of Michigan
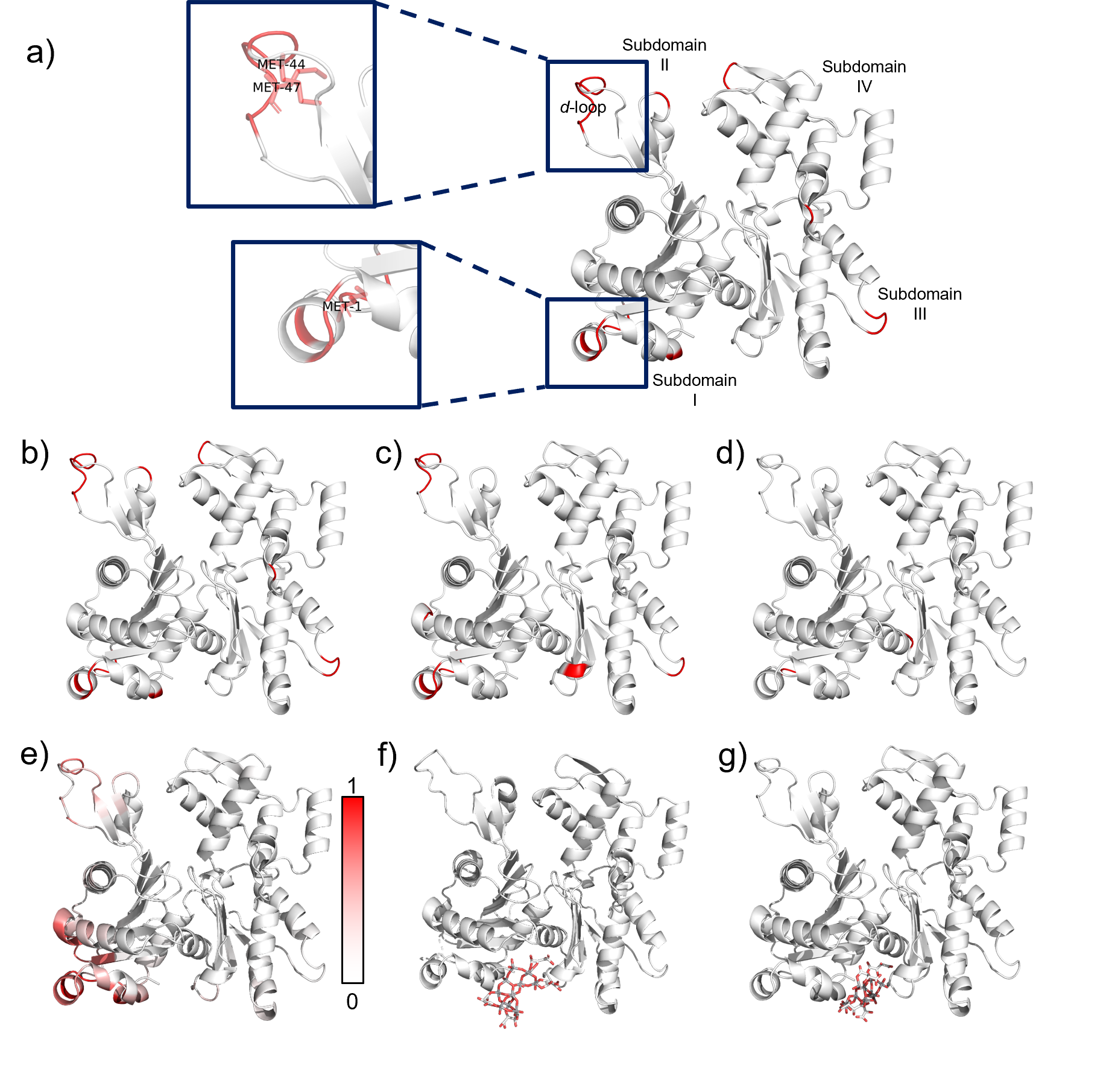
Since the rapid rise of nanotechnology as a field in the second half of the twentieth century, nanoparticles (NPs) and nanostructured materials have been examined for many different biological applications, such as diagnostics, drug delivery, and antimicrobial applications1. Just as NP properties differ from that of their bulk material, NPs interact with proteins differently than a flat surface of their bulk material due to the high proportion of surface atoms and the high curvature of the surface.9 Biomimetic chiral nanomaterials self-assembled from chiral templates have demonstrated potential in biological applications2. The chirality of NPs influences how NPs and nano-structured surfaces interact with these proteins and consequentially, how they affect biological systems3. Modeling and predictions the interactions between proteins and NPs can help us design NPs with greater selectivity and efficacy for different applications. However, chiral nanoparticles and nanostructures often have complex geometries and multiscale chirality, making them difficult to model. Currently, protein interactions with chiral nanoparticles are modeled with highly sophisticated molecular dynamics (MD) simulations, but these are computationally intensive, meaning that they are often limited to NPs with simple geometry and short time scales of several hundred nanoseconds4. Here, we explore how we can adapt existing methods to protein-nanoparticle applications and ways to address the challenges of predicting chiral protein-nanoparticle interactions.