2025 AIChE Annual Meeting
(276d) Next-Generation Peptide Sequencing Via Chemoenzymatic Reverse Translation of Peptides into DNA
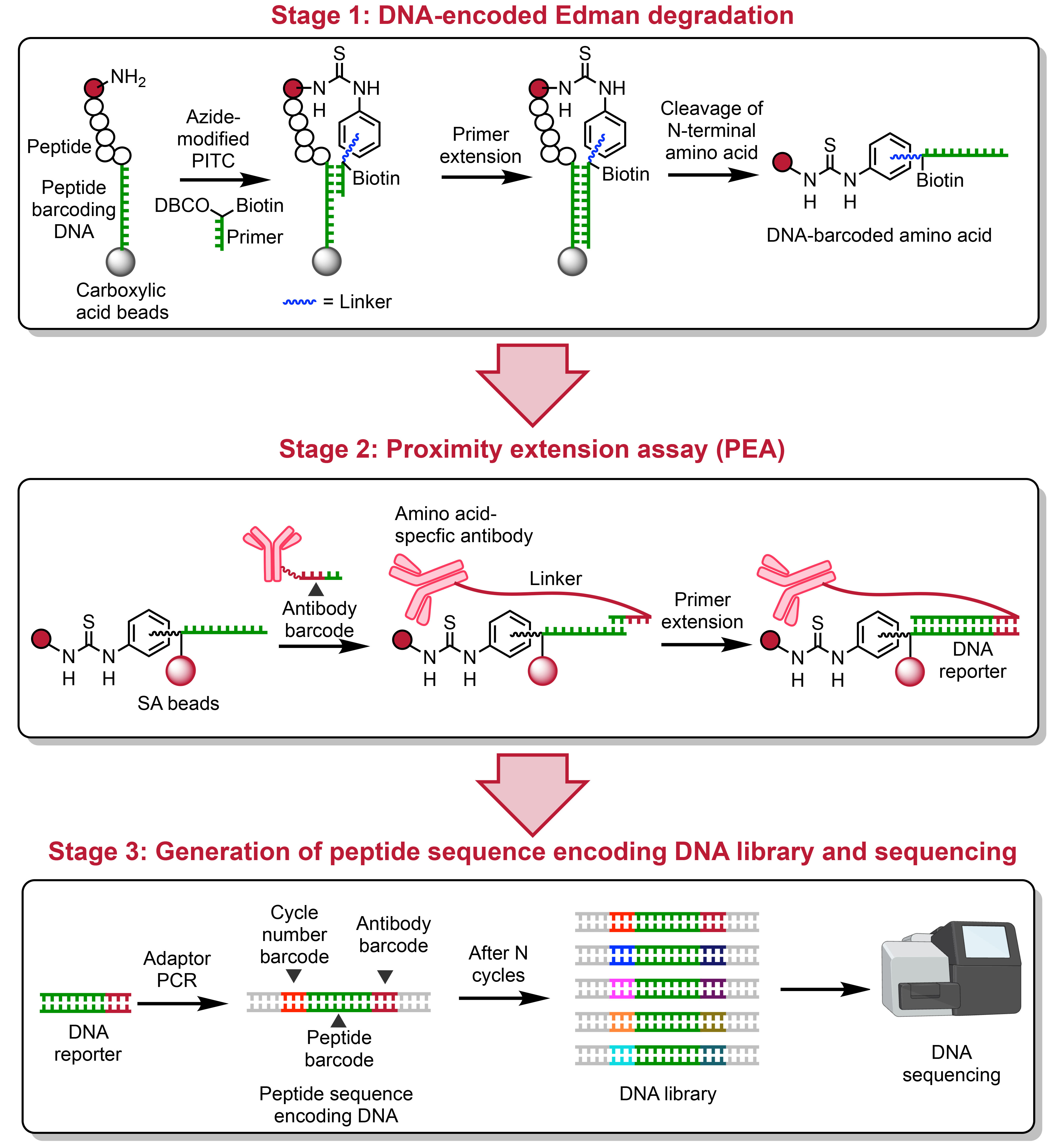
Proteins carry out the majority of cellular functions. While single-cell transcriptomics provides rich information on the expression level of proteins, it does not reveal the entire cellular proteome due to transcriptional regulation and post-translational modifications. As a result, the consequences of changes in expression can only be fully understood by assessing the protein content of a cell. This has spurred considerable interest in the development of technologies that enable single-molecule analysis and quantitation of proteins. Although important headway is being made regarding single-molecule protein profiling and quantitation, there is still an unmet need for methods that can accurately identify a wide range of proteins—including PTM variants—at a meaningful throughput. Here, I will present the development of peptide sequencing via chemoenzymatic reverse translation of peptide sequences into barcoded DNA libraries that encode the sequence of peptides. This approach, for the first time, enables peptide sequence information to be amplified by PCR and analyzed via high-throughput DNA sequencing. This method allows the detection of peptides in a mixture at the sub-zeptomole level. With further refinement and development, this approach will be amenable to the massively parallel profiling of both unmodified and PTM-bearing proteins, opening up exciting new opportunities for single-molecule proteomics.