2025 AIChE Annual Meeting
(537g) Molecular Motor Thermodynamics from Theory and Simulation
Author
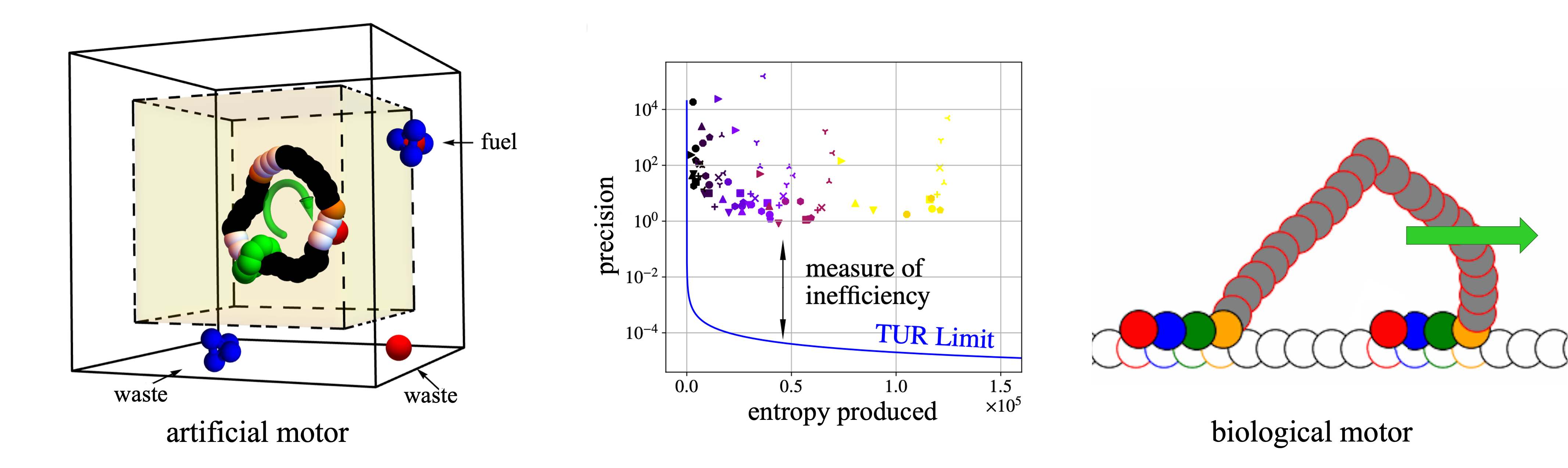
Molecular motors convert chemical energy into mechanical work at the nanoscale, extracting energy from a fuel to waste reaction. Like their macroscopic counterparts, they have a maximum thermodynamic efficiency but unlike large-scale machines, they experience significant thermal fluctuations that can cause backward steps. In this context, precision, the variance of forward versus backward motion, becomes an important performance metric. Recent theoretical developments known as thermodynamic uncertainty relationships (TURs) provide bounds on precision in terms of entropy production. To achieve high precision, a motor must generate a large amount of entropy.
In this talk, I will present simulation methodologies and coarse-grained models for simple artificial molecular motors. I will demonstrate how to extract entropy production from these simulations and apply TURs to quantify how closely motors approach fundamental thermodynamic limits. I will then extend this approach to biological motor proteins, focusing on myosin, a two-headed motor that walks hand-over-hand along actin filaments and plays a central role in muscle contraction and intracellular transport. Our coarse-grained model enables detailed quantification of myosin’s stepping mechanism and thermodynamics. Illuminating the principles that govern biological motors could guide the design of more effective artificial molecular machines.