2025 AIChE Annual Meeting
(138e) Metabolic Modeling of Chicken Cells for Cultivated Meat: Reconstruction of a High-Quality Genome-Scale Network
Authors
Chicken is one of the most widely consumed meats globally and a prime candidate for cultivated meat technologies. However, computational tools to support chicken-specific media design are lacking. The existing genome-scale metabolic model for Gallus gallus, iES1300, exhibits critical limitations, including extensive stoichiometric inconsistencies, mass and charge imbalances, and a high proportion (63%) of universally blocked reactions. Additionally, 35% of the metabolites in iES1300 are either dead-end or orphan, some with unrealistic charges, rendering the model inadequate for downstream metabolic analyses.
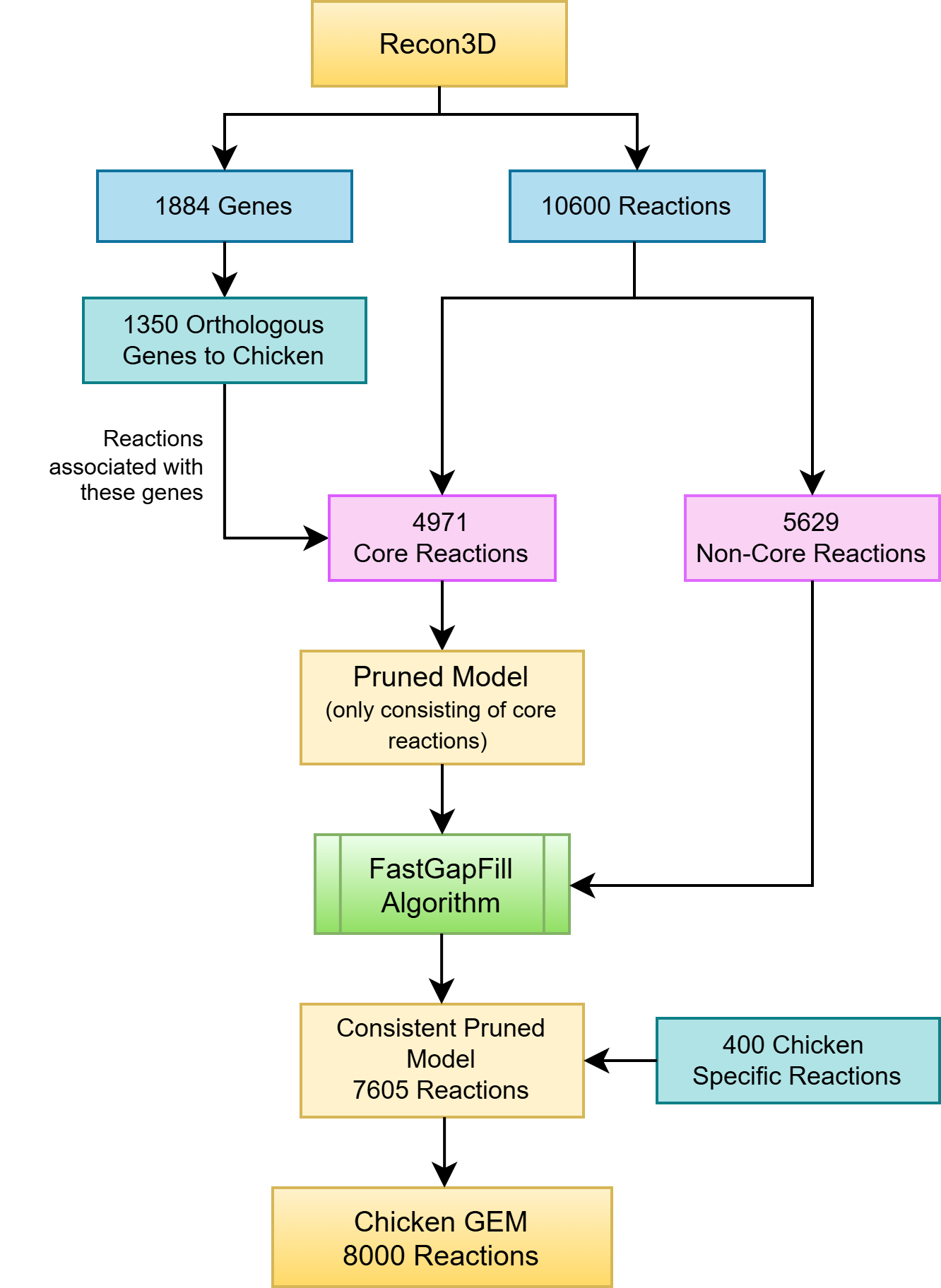
To address these limitations, we developed a refined and compartmentalized eukaryotic GEM for chicken using an orthology-based reconstruction approach. We leveraged Recon3D, the most recent flux-consistent and comprehensive version of the human metabolic network, as the primary reference due to its high metabolic coverage and absence of blocked reactions or dead-end metabolites. We replaced the human genes in Gene-Protein-Reaction (GPR) associations with 1,350 chicken orthologues identified from 1,884 human genes in Recon3D using the Genome Pair View tool. All chicken gene-associated reactions were defined as core reactions, while the remaining reactions were treated as non-core. To ensure metabolic completeness, we performed gap-filling using the FastGapFill function by PSAMM Tools, identifying the minimal set of non-core reactions necessary to support flux over core pathways.
Without independent identification of chicken-specific metabolic functions, the reconstructed model would risk being merely a pruned version of the human reference network. To address this, we systematically integrated metabolic pathway data from KEGG and other biochemical databases and literature sources to ensure species-specific metabolic coverage. The final GEM comprises over 5,800 metabolites and up to 8,000 reactions, with improved metabolite and reaction annotations, complete pathway connectivity, and no dead-end or orphan metabolites. This network can be used for metabolc flux simulations with extracellular metabolite profiles obtained from spent media analysis, providing insights on nutrient and growth factor utilization and guiding rational optimization of media composition. Beyond cultivated meat applications, this refined GEM also provides a robust framework for studying avian metabolism and advancing metabolic engineering and systems biology research in poultry science.