2025 AIChE Annual Meeting
(588h) Machine Learning–Driven Optimization of Ionizable Lipids for High-Efficiency mRNA Delivery to Liver and Gene Editing in the Lung Epithelia
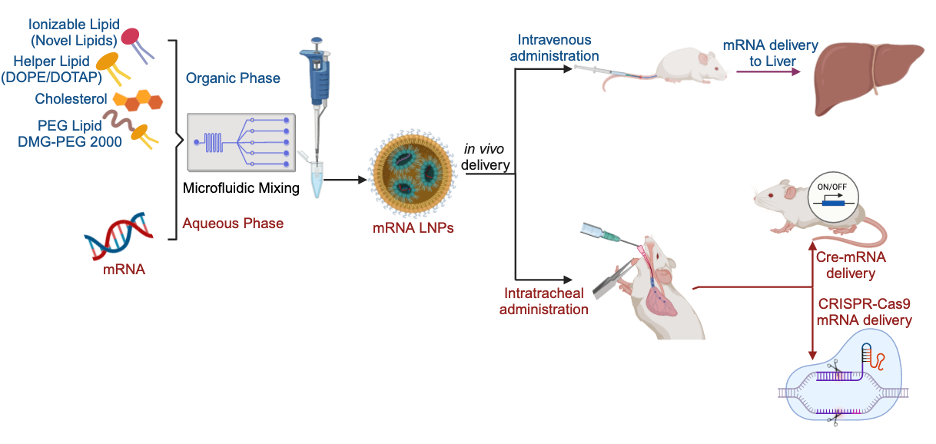
Messenger RNA, or mRNA, delivered via lipid nanoparticles or LNPs has emerged as a transformative platform for a wide range of therapeutic applications, from protein replacement to correcting genetic disorders. The clinical success of the Pfizer-BioNTech and Moderna COVID-19 vaccines truly brought this technology into the spotlight. Building on that foundation, our research aims to develop next-generation mRNA delivery systems to treat chronic diseases—particularly those affecting the liver and lungs, such as Alpha-1 antitrypsin deficiency and cystic fibrosis. We started by targeting liver delivery. To do this, we synthesized a library of 384 novel ester-derived biodegradable ionizable lipids. To minimize animal use, especially C57BL/6 mice, we developed a high-throughput in vivo screening platform using peptide-barcoded mRNA. This allowed us to test LNP formulations systematically. From 24 different formulations, we identified one that achieved a threefold increase in liver transfection efficiency compared to our original benchmark. We then validated these findings using a human erythropoietin (hEPO) expression assay. Impressively, our lead biodegradable lipid delivered mRNA to the liver 4.5 times more effectively than DLin-MC3-DMA—a clinically validated standard. To push the efficiency even further, we introduced a deep learning–based platform we call LiON—short for Lipid Optimization using Neural Networks. Trained on a dataset of over 9,000 mRNA delivery outcomes from both in vitro and in vivo studies, LiON could accurately predict top-performing lipids. It identified lipids like RJ-A30-T01, which not only outperformed existing clinical lipids like MC3 and SM-102, but also validated the strength of AI-guided design in this space. Shifting focus to lung delivery, we sought to enable CRISPR-Cas9–based gene editing in the lungs. We designed a three-component delivery system and synthesized more than 700 biodegradable lipids. Using Cre-recombinase mRNA in Ai9 tdTomato reporter mice, we achieved up to 53% tdTomato-positive cells—double what we previously saw with nebulized Cre-mRNA. Finally, RCB-4-8 LNPs, when loaded with Cas9 mRNA/sgRNA at low and high doses and quantified gene-editing, could result in 3 and 7% tdTomato+ cells, respectively. These Cas9 mRNA/sgRNA loaded mRNA were found to gene editing in both club cells and ciliated cells. Application of these CRISPR-Cas9 mRNA for gene/cancer therapy by removing faulty genes or correcting mutations is ongoing.