2025 AIChE Annual Meeting
(394af) LLM-Powered Knowledge Graph Framework - the Analysis of Biochemical Networks
Authors
The pipeline begins with the parsing of SBML-encoded GSMs to extract reaction stoichiometries, gene–protein–reaction (GPR) associations, metabolite roles, and compartmentalization. These entities are mapped to our custom meta ontology and structured as RDF triples for ingestion into the KG. The current implementation encodes explicit biochemical and topological relationships, including enzyme-catalyzed reactions, substrate–product flows, and regulatory constraints. Future versions of the framework will incorporate learned vector embeddings—generated via graph-based and language model-based techniques—to represent metabolic components in a continuous latent space. These embeddings will facilitate semantic similarity search, entity disambiguation, and relevance scoring within a retrieval-augmented generation (RAG) architecture, improving both the precision and interpretability of system responses.
At present, the system uses general-purpose, pre-trained LLMs such as Llama3 and CodeLlama in a zero-shot configuration. These models are integrated via an inference API layer to process user prompts, guide the SPARQL query generation, and generate explanatory responses from the structured KG outputs. While no fine-tuning has yet been performed, we anticipate that domain adaptation—via continual pretraining or supervised fine-tuning on corpora from sources such as BioCyc, KEGG, and EcoCyc—will significantly enhance biological reasoning capabilities. In future iterations, fine-tuning will be paired with advanced prompting strategies, including retrieval-conditioned prompts, chain-of-thought (CoT) generation, and dynamic prompt templating based on KG context.
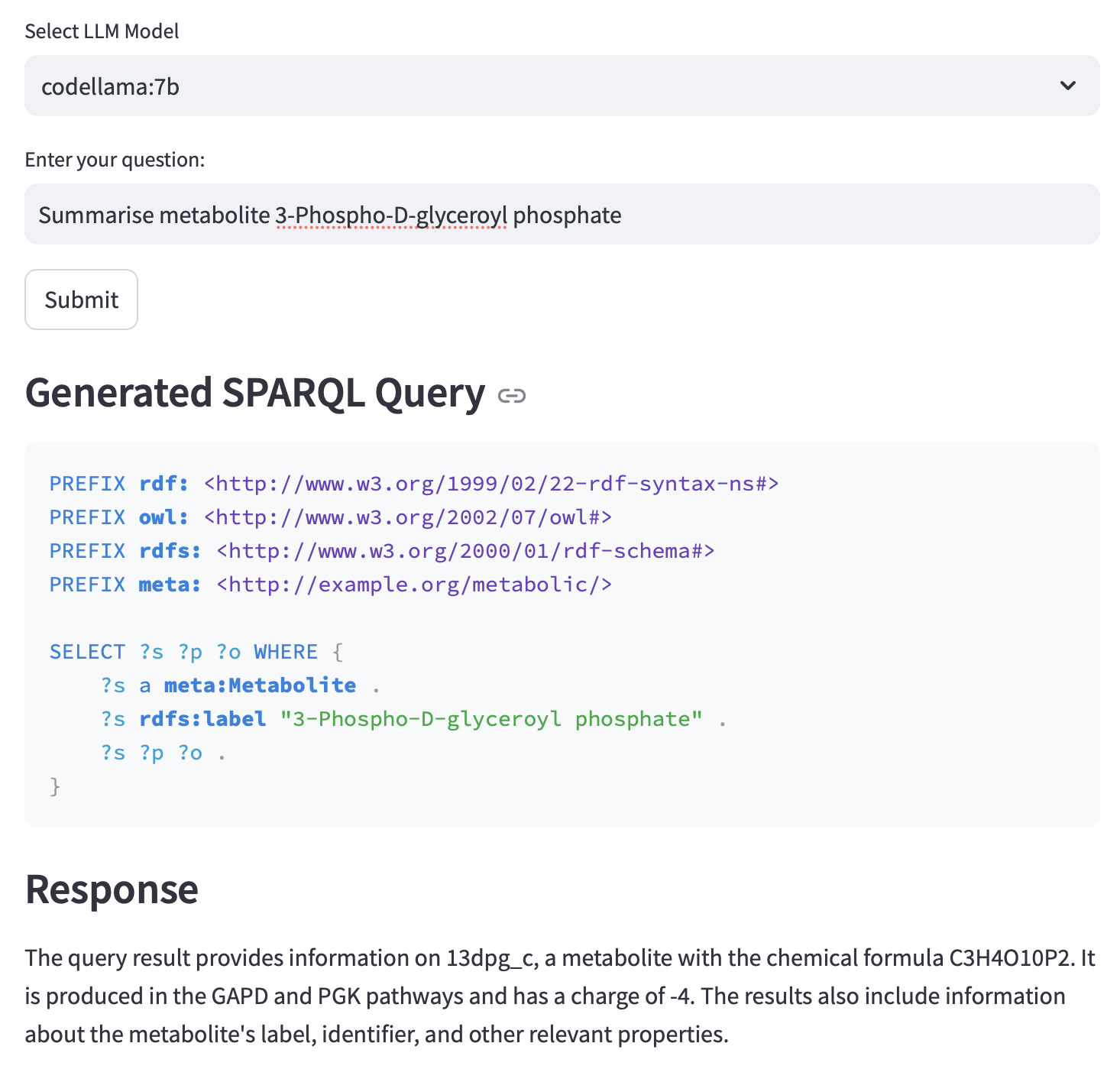
User interaction is mediated through a multi-stage pipeline: natural language queries are parsed and interpreted by the LLM, which then produces formal SPARQL queries targeting relevant entities and relations within the KG. The resulting query outputs are processed and distilled into biologically coherent responses, rendered in natural language and complemented by interactive network visualizations. Visual layers include reaction-centric views, metabolite-centric subnetworks, and pathway-level abstractions, each driven by user-defined filters or constraints.
This architecture supports a new paradigm for exploratory metabolic modeling, offering a human-in-the-loop interface for interrogating complex biological systems without requiring deep familiarity with query languages or formal model structures. By coupling symbolic representations (via RDF and ontologies) with the generative and interpretive capabilities of LLMs, the system enables a hybrid reasoning environment that supports hypothesis generation, data interpretation, and model curation.
Ongoing and future development efforts include the benchmarking of LLM-generated queries and responses against ground-truth pathway annotations and expert-curated outputs, extension of the framework to incorporate regulatory and signaling models beyond GSMs, optimization of KG query latency for real-time interaction, and integration of quantitative modeling layers including flux balance analysis (FBA) and kinetic rate laws. Additionally, embedding-based indexing will be introduced to support semantic search, enabling the LLM to retrieve and rank contextually relevant subgraphs to support high-fidelity RAG workflows. Together, these enhancements aim to establish a biologically grounded, computationally scalable platform for LLM-assisted metabolic network analysis.