2025 AIChE Annual Meeting
(392an) Latent Space-Based Bayesian Optimization for Efficient Screening of Enzyme Mutants in Biocatalysis
Authors
Protein engineering is essential for modern biocatalysis, both using rational methods and artificial intelligence (AI) designs. However, the vast sequence space of enzyme variants presents a significant challenge when screening protein libraries. Traditional screening methods are costly and time-intensive, often requiring several weeks of experimental validation. This bottleneck is particularly limiting for AI-driven enzyme design, where it is not possible to test all computationally generated variants in a library. To accelerate discovery of optimized enzymes in biocatalysis, an efficient strategy is needed to navigate the sequence space while minimizing experimental costs.
Approach: Latent space Bayesian optimization with mechanistic insights
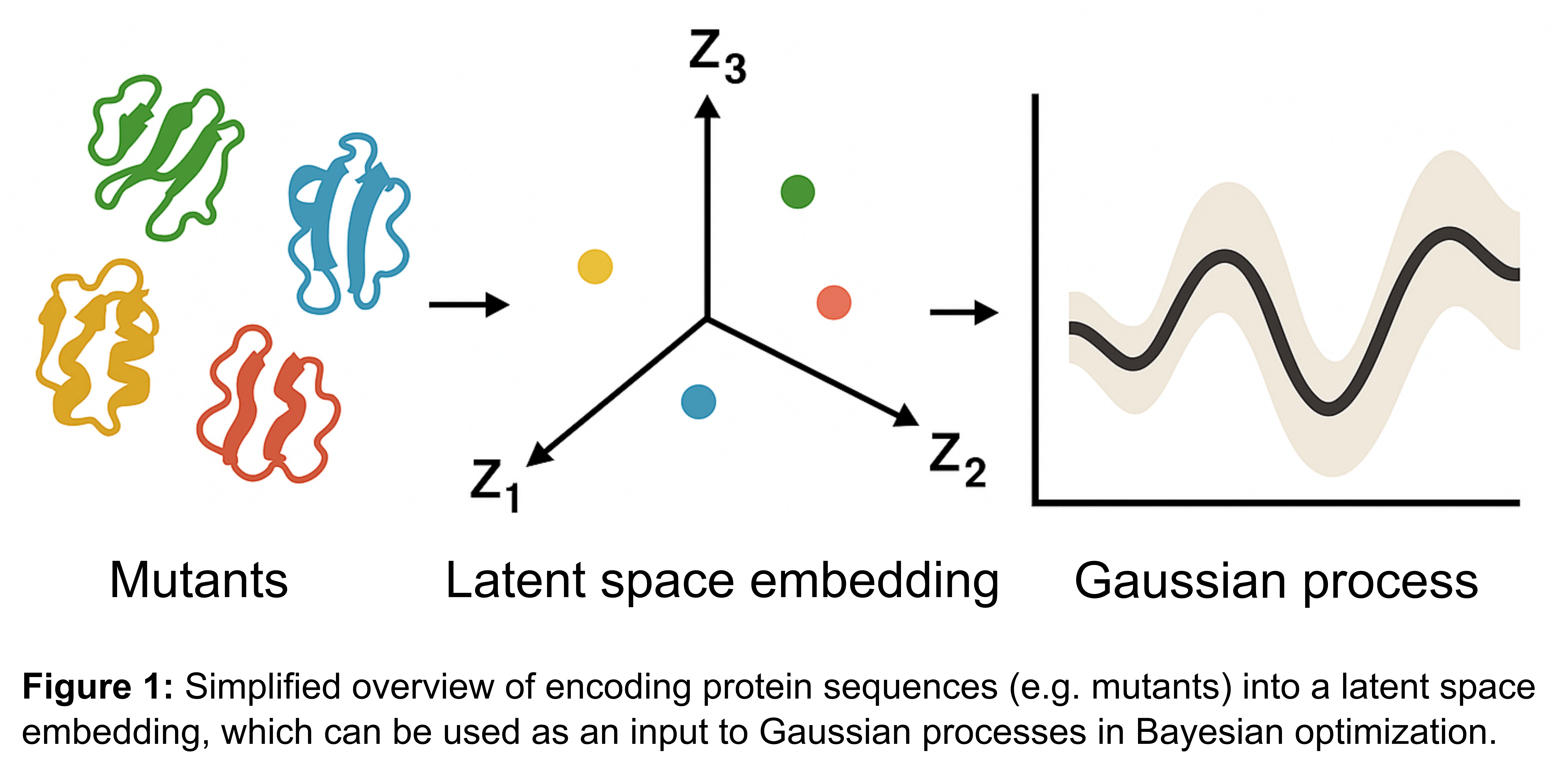
Bayesian optimization (BO) is a powerful tool for guiding expensive experimental searches, using surrogate models like Gaussian processes (GPs) to model complex and unknown objective functions. However, directly applying BO to high-dimensional protein sequences is challenging. To address this, we employ BO in a low-dimensional latent space, created using embeddings from deep learning models, such as variational autoencoders and protein language models like ProtT5 [1] and ESM-2 [2]. These embeddings enable a structured representation of the sequence space, facilitating efficient search through the sequence landscape (Figure 1).
Additionally, we integrate a mechanistic model for the reaction kinetics of our case study, which accounts for substrate inhibition. While the mechanistic model provides valuable domain knowledge to extract kinetic parameters, it cannot capture the complex relationships between sequence variations and enzyme activity. Here, BO plays a crucial role in navigating these intricate relationships, optimizing for high-performance enzyme variants by incorporating both data-driven insights and mechanistic constraints.
Application: Optimizing cGAS mutants for enhanced cGAMP production
We apply this framework to optimize cyclic GMP-AMP synthase (cGAS), a key enzyme in immune signaling with applications in autoimmune disease and cancer therapy [3]. It has already been shown that this enzyme has promising properties as a biocatalyst. Importantly, the bioprocess for the synthesis of cyclic dinucleotides, compounds with antiviral and anti-cancer properties, is more environmentally friendly than the respective chemical process [4,5].
In this work, BO is used to identify promising candidates for improved cyclic guanosine monophosphate–adenosine monophosphate (cGAMP) production from a mutant library with 100 variants of cGAS. As a full screening of all candidates is impractical, a strategy is needed to select small subsets (2-4 candidates per batch) for manual screening. Here, BO-guided screening efficiently prioritizes high-performing mutants, reducing the number of required experimental evaluations compared to random or exhaustive searches. The suggested variants of the batch BO process will be validated in wet-lab experiments to identify the best cGAS mutant in the library regarding stability and activity.
Outlook: Industrial relevance and future applications
Efficient mutant screening is essential for advancing biocatalysis and protein engineering. By integrating BO with advanced sequence embeddings, our approach accelerates the identification of optimized variants while reducing experimental costs. While demonstrated with the enzyme cGAS, the approach is widely applicable to various enzymes and protein engineering tasks. Future work will focus on refining sequence representations and exploring novel GP kernel functions to enhance predictive accuracy. Ultimately, integrating this framework with automated experimentation could significantly accelerate AI-driven enzyme engineering and industrial biocatalyst development.
[1] Elnaggar, Ahmed, et al. "ProtTrans: towards cracking the language of life’s code through self-supervised learning." IEEE Transactions on Pattern Analysis and Machine Intelligence 44 (2021): 7112-7127.
[2] Lin, Zeming, et al. "Evolutionary-scale prediction of atomic-level protein structure with a language model." Science 379.6637 (2023): 1123-1130.
[3] Ablasser, Andrea, Chen, Zhijan. "cGAS in action: Expanding roles in immunity and inflammation." Science 363 (2019): 1055.
[4] Rosenthal, Katrin, et al. "Catalytic Promiscuity of cGAS: A Facile Enzymatic Synthesis of 2′-3′-Linked Cyclic Dinucleotides." ChemBioChem 21 (2020): 3225-3228.
[5] Becker, Martin, et al. "Comparative Life Cycle Assessment of Chemical and Biocatalytic 2’3’-Cyclic GMP-AMP Synthesis." ChemSusChem 16 (2023): e202201629.