2025 AIChE Annual Meeting
(543g) Invited Talk: Improving CRISPR-Cas9 Homology Directed Repair in Ial-PiD2 Insect Cells from Plodia Interpunctella
Authors
Plodia interpunctella has a well-documented history in pest management as a significant global pest that affects stored commodities and processed food products 13. More recently, it has emerged as a candidate for silk-based biomaterial production 14-16. To validate and optimize CRISPR/Cas9 HDR in this system, the fibroin-heavy chain (Fib-H) gene was selected as the initial target 17. Fib-H is the primary structural component of Lepidopteran silk and a commercially significant biopolymer. While its amino acid composition and structure vary across species, it remains the foundation of silk fiber formation and is the key protein used in silk-based biomaterials. Most silk-based biomaterials18 are derived from fibroin proteins extracted from the domesticated silkworm, Bombyx mori; however, sourcing or modifying fibroins from alternative species could expand the range of material properties, offering new possibilities for medical and commercial applications.
In this study, several factors influencing CRISPR/Cas9 HDR knock-in efficiency were examined. First, the ratio of Cas9 to sgRNA plays a critical role in the formation of the ribonucleoprotein (RNP) complex, directly affecting target site recognition and cleavage efficiency. Additionally, the amount of donor DNA template can impact the availability of homologous sequences for repair, influencing integration success. To further enhance HDR, cell cycle synchronization was enhanced using 2-hydroxyurea, which arrests cells in the G0/G1 phase, allowing for delivery of the CRISPR/cas9 and sgRNA when homology-directed repair is most active. By optimizing these parameters, we improved targeted integration efficiency in P. interpunctella, IAL-PiD2, cells. These efforts are a critical step for using the IAL-PID2 cell line for future therapeutic protein production.
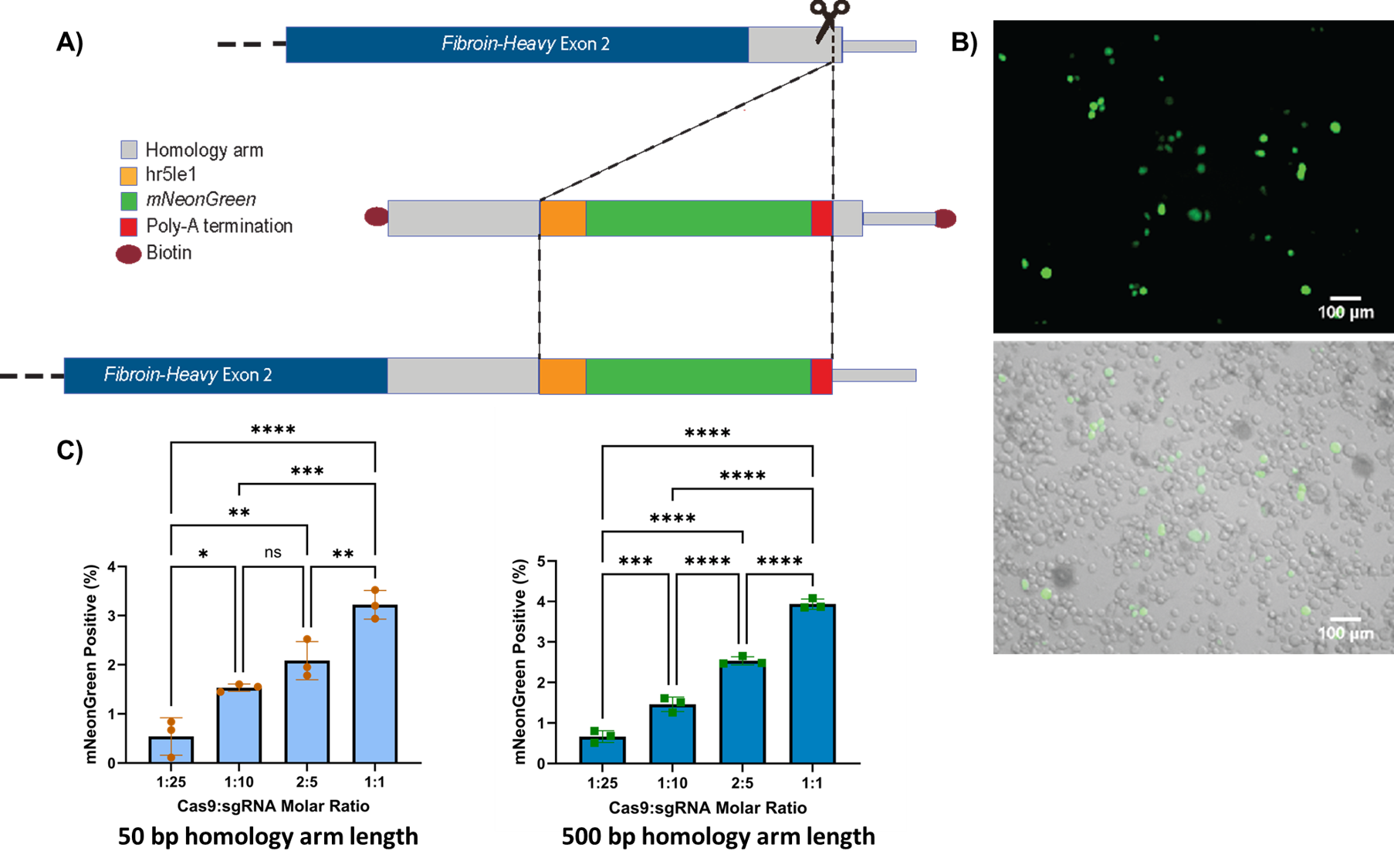
Figure 1 shows the general modification plan for the homology directed repair in the Fib-H gene, example images of modified IAL-PID2 cells expressing mNeonGreen, and the comparisons between cas9 and sgRNA concentrations for two homology arm lengths, supporting these results.
References
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nature biotechnology. 2015;33(5):543-8.
- He X, Tan C, Wang F, Wang Y, Zhou R, Cui D, You W, Zhao H, Ren J, Feng B. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic acids research. 2016;44(9):e85-e.
- Di Stazio M, Foschi N, Athanasakis E, Gasparini P, d’Adamo AP. Systematic analysis of factors that improve homologous direct repair (HDR) efficiency in CRISPR/Cas9 technique. PLOS ONE. 2021;16(3):e0247603. doi: 10.1371/journal.pone.0247603.
- Hayashi A, Tanaka K. Short-homology-mediated CRISPR/Cas9-based method for genome editing in fission yeast. G3: Genes, Genomes, Genetics. 2019;9(4):1153-63.
- Singh S, Banerjee A, Vanden Broeck A, Klinge S. Rapid clonal identification of biallelic CRISPR/Cas9 knock-ins using SNEAK PEEC. Scientific Reports. 2023;13(1):1719.
- Martin RM, Ikeda K, Cromer MK, Uchida N, Nishimura T, Romano R, Tong AJ, Lemgart VT, Camarena J, Pavel-Dinu M. Highly efficient and marker-free genome editing of human pluripotent stem cells by CRISPR-Cas9 RNP and AAV6 donor-mediated homologous recombination. Cell stem cell. 2019;24(5):821-8. e5.
- Nakata M, Ueno M, Kikuchi Y, Iwami M, Takayanagi-Kiya S, Kiya T. CRISPR/Cas9-and Single-Stranded ODN-Mediated Knock-In in Silkworm Bombyx mori. Zoological science. 2024;41(6).
- Chenouard V, Leray I, Tesson L, Remy S, Allan A, Archer D, Caulder A, Fortun A, Bernardeau K, Cherifi Y. Excess of guide RNA reduces knockin efficiency and drastically increases on-target large deletions. Iscience. 2023;26(4).
- Zhang X-H, Tee LY, Wang X-G, Huang Q-S, Yang S-H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Molecular therapy Nucleic acids. 2015;4.
- Zhang J-P, Li X-L, Li G-H, Chen W, Arakaki C, Botimer GD, Baylink D, Zhang L, Wen W, Fu Y-W. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome biology. 2017;18:1-18.
- Zhu L, Mon H, Xu J, Lee JM, Kusakabe T. CRISPR/Cas9-mediated knockout of factors in non-homologous end joining pathway enhances gene targeting in silkworm cells. Scientific reports. 2015;5(1):1-13.
- Lynn D, Oberlander H. The establishment of cell lines from imaginal wing discs of Spodoptera frugiperda and Plodia interpunctella. Journal of Insect Physiology. 1983;29(7):591-6.
- Mohandass S, Arthur F, Zhu K, Throne JE. Biology and management of Plodia interpunctella (Lepidoptera: Pyralidae) in stored products. Journal of Stored Products Research. 2007;43(3):302-11.
- Shirk BD, Torres Pereira Meriade Duarte I, McTyer JB, Eccles LE, Lateef AH, Shirk PD, Stoppel WL. Harvesting Silk Fibers from Plodia interpunctella: Role of Environmental Rearing Conditions in Fiber Production and Properties. ACS Biomaterials Science & Engineering. 2024.
- Eccles LE, Aikman EL, McTyer JB, Cruz ILM, Richgels AL, Stoppel WL. Exploring the functional properties of Plodia interpunctella silk fibers as a natural biopolymer for biomaterial applications. Materials Today Communications. 2025;42:111416.
- Nikodijević D, Ćurčić Milutinović M, Radenkovic N, Blagojević S, Vasiljević A, Jurisic V, Predojević D, Vukajlović F, Pešić S. Silk of the Indian meal moth Plodia interpunctella (Hübner, 1813) affects the human colon cancer cells. Kragujevac Journal of Science. 2024.
- Kawahara AY, Storer CG, Markee A, Heckenhauer J, Powell A, Plotkin D, Hotaling S, Cleland TP, Dikow RB, Dikow T. Long-read HiFi sequencing correctly assembles repetitive heavy fibroin silk genes in new moth and caddisfly genomes. Gigabyte. 2022;2022.
- Holland C, Numata K, Rnjak-Kovacina J, Seib FP. The Biomedical Use of Silk: Past, Present, Future. Adv Healthc Mater. 2019;8(1):e1800465. Epub 20180920. doi: 10.1002/adhm.201800465. PubMed PMID: 30238637.