2025 AIChE Annual Meeting
(687a) Invited Talk: Generating a High-Throughput Platform Technology for the Discovery, Assembly, and Evaluation of Immune-Modulating Nanobodies
Toward this, we will share three stories from our group that introduce a novel end‑to‑end workflow that couples complementary platform technologies: (1) development of a universalized surface-receptor for yeast surface-display, offering an approach to develop and screen for binders towards proteins that are traditionally challenging to express in yeast and at specific sites of interest on the protein, (2) RADAR (Rapid Agonist and antagonist Discovery for dimeric-activated immune Receptors) as a novel functional yeast-surface display screening technology to directly measure nanobody-mediated receptor activation via split-fluorescent protein reporting and HA-tagged binding quantification, and (3) a modular click‑chemistry assembly system based on engineered immune-modulating nanobodies for plug‑and‑play construction of bi‑ and tri‑specific nanobody‑based immune cell engagers.
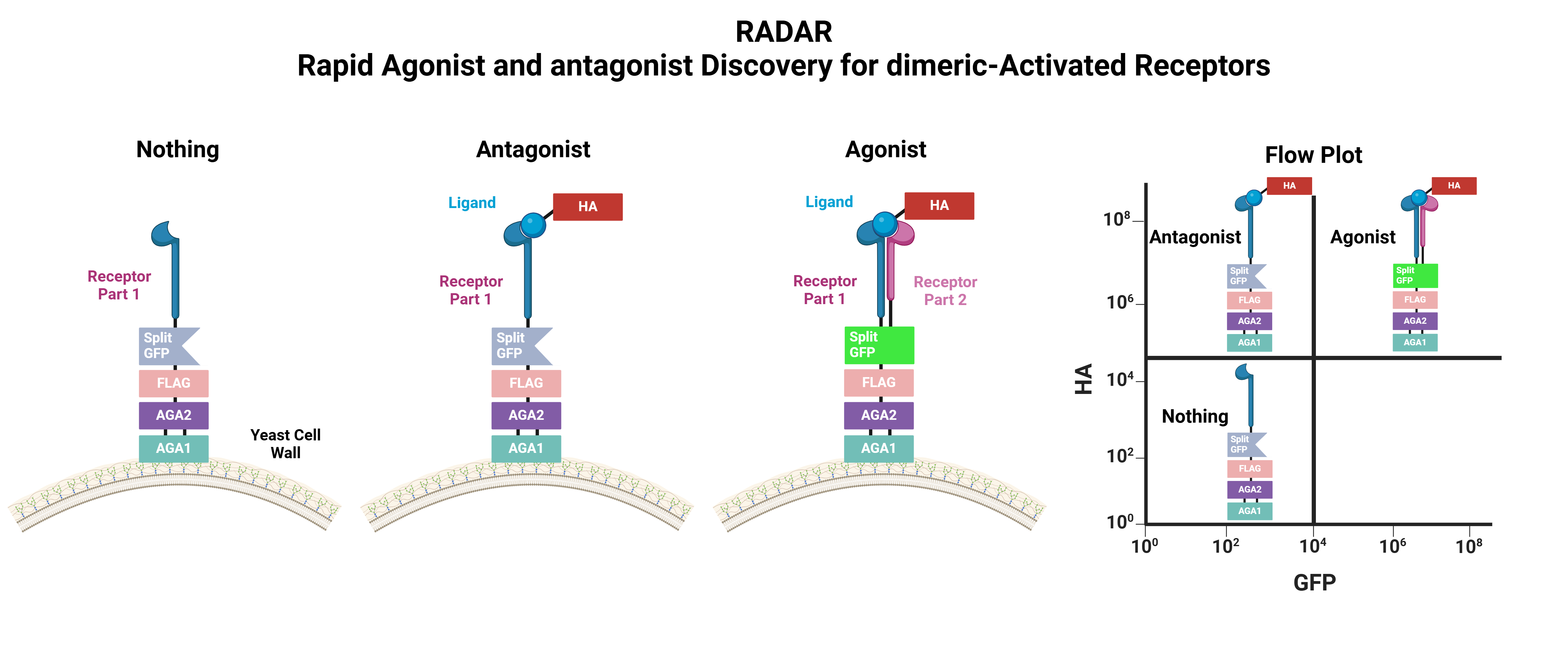
In our talk, Ethan Slaton (PhD Candidate) will tell the story of how RADAR uses split‑fluorescent protein fusions on interleukin receptor monomers to quantify ligand‑induced dimerization and consequent signaling – offering simultaneous selection for binding affinity and functional outcome. In proof‑of‑concept screens targeting an interleukin receptor, we isolated a library of nanobodies exhibiting distinct agonist or antagonist behaviors, reducing lead discovery to under four weeks.
Using engineered proteins, Roman Adomanis (PhD Candidate) will showcase our click‑chemistry bioconjugation platform that uses ncAA‑installed sites in the backbone of nanobody domains and hetero‑functional polymer linkers to assemble a panel of immune-modulating multidomain constructs, without requiring genetic fusion. This approach overcomes traditional protein design and synthesis limitations related to enzymatic or chemical oligomerization, offering rapid combinatorial generation of diverse bispecific and trispecific formats to drive potent and synergistic tumor killing to generate anti-tumor immunity.
Combining these technologies offers a novel pipeline to (1) rapidly design potential nanobodies as binders to traditionally complex, toxic, and challenging receptors in yeast, (2) screen the engineered nanobody for downstream immune responses, and (3) build modular assembly toolboxes of multidomain nanobodies for rapid preclinical testing of protein-protein interactions that generate lasting and robust anti-tumor immunity. Here, we share our recent strategies that provide insight into the development of a versatile, accelerated pipeline – from computational design and library screening through rapid multispecific construct generation – to design, build, test, and learn about next‑generation nanobody‑based immunotherapeutics for precision cancer treatment.