2025 AIChE Annual Meeting
(482g) Invited Talk: Advancing Biocatalysis on Bacterial Spores through an Extensive Survey of Coat Components
Author
Nikhil U. Nair - Presenter, 5/7/2018
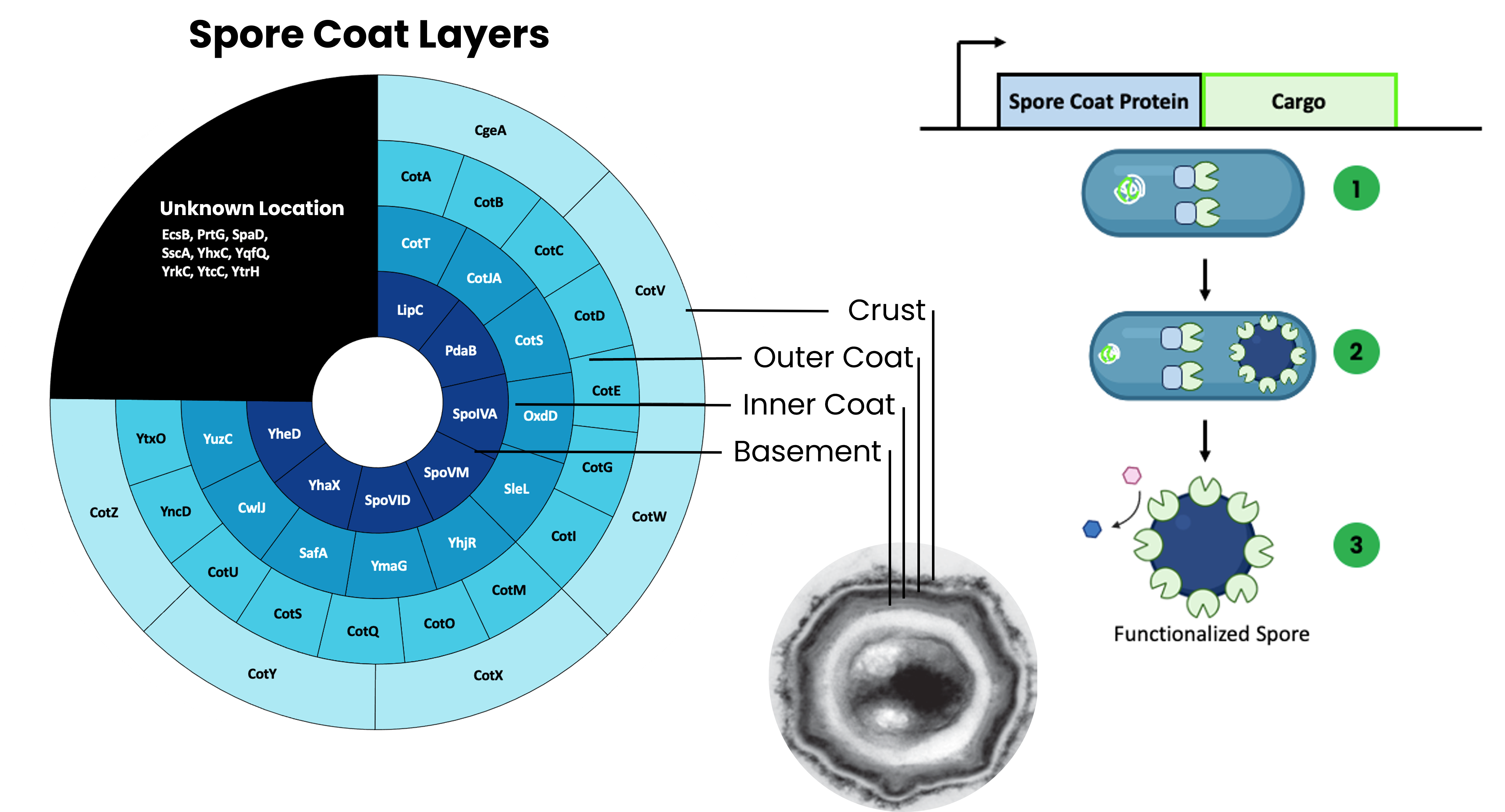
Spore display is finding increasing use in a variety of applications such as cell-free metabolic engineering, enzymatic biocatalysis, vaccine delivery, environmental remediation, etc. for several reasons. Since spores are inherently highly robust to thermal, pH, proteinase, and solvent stresses, they often confer some of these properties to anchored proteins as well. Further, they are relatively unique among biological surface-display systems since they are assembled within the cytoplasm prior to release, and thus, proteins do not need to be secreted to be loaded and surface available. Finally, spores derived from Bacillus subtilis can be rapidly engineered since the microbe is naturally competent. Despite its promise, the potential of spore display has not yet been fully realized. Specifically, spore-display lacks a robust set of well-characterized biological parts like other microbial surface display technologies. While there are more than 50 known spore proteins, only 12 have been used for anchoring enzymes. In this study, we sought to thoroughly characterize the potential of B. subtilis spore coat proteins as fusion partners for spore-displayed enzymes. We compiled a list of 44 known spore coat proteins and designed 88 constructs to make N and C-terminal fusions to β-glucuronidase (GUS) from Escherichia coli to each potential anchor protein. Synthesis of these constructs was performed by the DOE JGI. Of these, only 52 could be synthesized and verified, representing 37 of the original 44 spore coat proteins. We then assessed the ability of each of these 37 spore coat proteins to act as functional fusion partners by quantifying enzyme activity, surface availability, and stability of the GUS fusions. We found a previously understudied protein as a highly promising candidate for spore display protein called SscA. Further characterization of the top candidates and combined use of multiple anchor proteins further enhanced the functional capacity of spores. Cumulatively, this study provides a comprehensive and systematic comparison of spore coat proteins for their potential as carriers of protein immobilization and builds fundamental, data-informed design principles for spore display. This work will expedite the adoption of spore-display as a genetically encoded protein immobilization technology for any number of applications.