2025 AIChE Annual Meeting
(207f) Hybrid Modeling of Industrial Bioreactors Using Physics-Informed Neural Networks (PINNs)
Authors
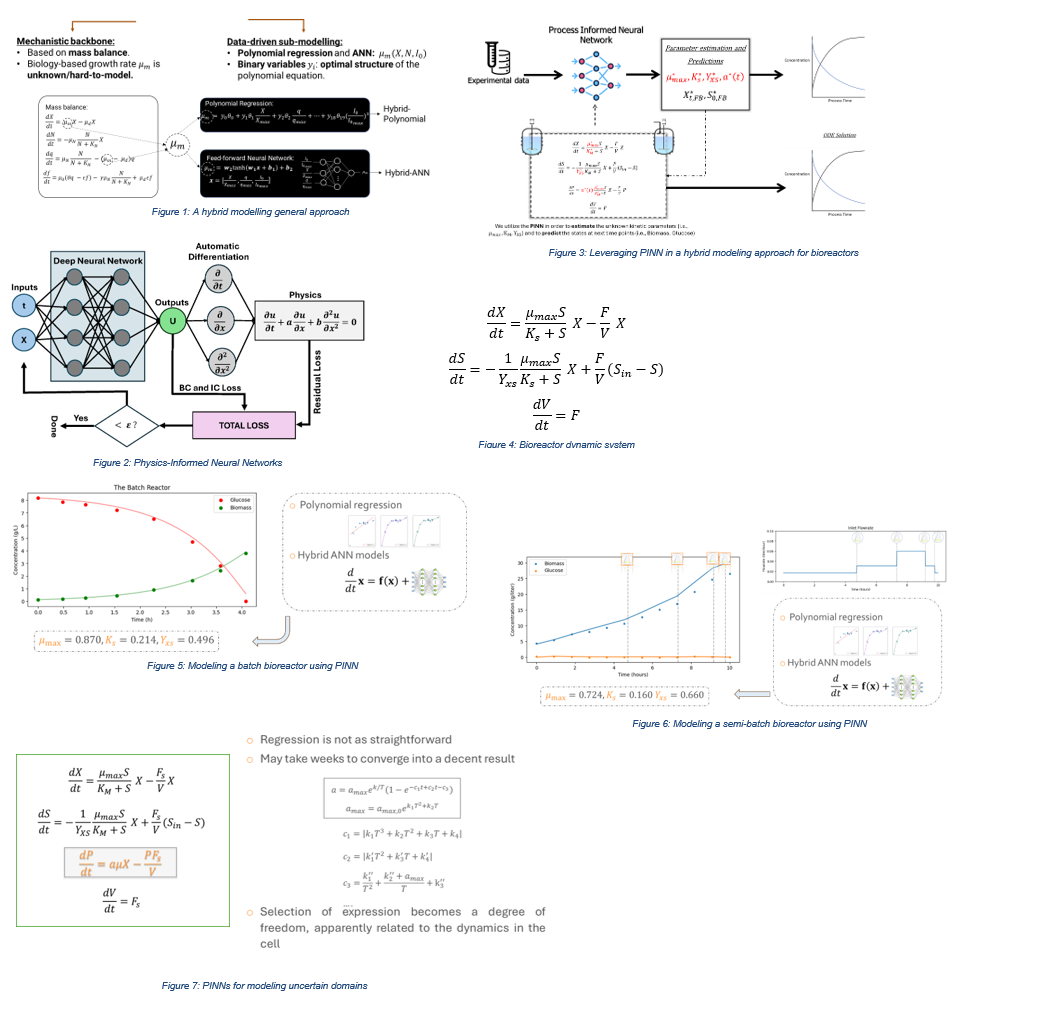
Physics-Informed Neural Networks (PINNs) (Figure 2) have emerged as a powerful approach for integrating first-principles knowledge with data-driven modeling. Unlike traditional neural networks that rely solely on data, PINNs incorporate physical laws—typically formulated as ordinary or partial differential equations—directly into the training process via the loss function. This fusion allows the model to learn not only from experimental measurements but also from the underlying scientific principles, enhancing generalization, interpretability, and performance, particularly in data-scarce settings. PINNs have already demonstrated success across a range of domains. By blending the strengths of machine learning with the rigor of mechanistic modeling, they offer a robust and physics-consistent alternative to conventional black-box approaches. PINNs are subsequently evaluated through an extensive series of experiments, including: (a) batch and semi-batch bioreactor models; and (b) scenarios where the underlying model structure is uncertain or partially unknown. In the first set of experiments, we demonstrated that PINNs can effectively capture state-space dynamics and deliver accurate short- and long-term state predictions for both batch and semi-batch bioreactor models (Figure 3).
We initially focused on assessing the ability of PINNs to dynamically learn the system’s kinetic parameters as the process evolves. Unlike traditional methods that require complete data trajectories for parameter estimation and model identification, the PINN-based approach enables continuous learning and real-time inference of unknown kinetic parameters. This adaptive modeling capability allows for more flexible and data-efficient analysis of bioprocesses, making PINNs particularly valuable in settings with sparse measurements or fluctuating operating conditions. Impressively, PINNs succeeded in reconstructing system states and dynamics with as little as two data points, demonstrating their resilience in low-data environments (Figure 5). This capability arises from their integration of governing physical principles into the training process, which allows them to extract meaningful insights even when observational data are minimal. In the semi-batch scenario, PINNs consistently maintained high accuracy even in the presence of abrupt shifts in the state space—such as sudden changes in inlet flow rate—where traditional models often struggle (Figure 6). Their ability to deliver accurate, real-time state estimation allows for timely adjustments to process conditions, facilitating more informed and proactive decision-making. Furthermore, as the process unfolds, the PINN continuously refines its representation of the kinetic parameter space, effectively addressing the limitations of static, fixed-parameter models. This adaptability is essential for achieving flexible, responsive, and efficient operation in the dynamic landscape of biomanufacturing.
The second phase of modeling targets the identification of unknown kinetics governing protein production. Candidate kinetic expressions are drawn from a curated set, informed by prior analyses of cellular behavior and encompassing a broad spectrum of dynamic profiles (Figure 7). As the bioprocess progresses, the PINN autonomously determines the most suitable kinetic model while concurrently optimizing its parameters. This model selection process is guided by a clearly defined criterion based on the PINN’s training loss, ensuring alignment with both data accuracy and underlying physical principles. The method reliably identifies the correct kinetic expression for previously uncharacterized protein production dynamics. This capability is particularly valuable in bioprocess development, where mechanistic insight is often incomplete. Accurately uncovering the governing kinetics not only strengthens process modeling but also enhances optimization strategies and accelerates development timelines.
This study underscores the significant potential of Physics-Informed Neural Networks (PINNs) as hybrid modeling frameworks for industrial bioreactor applications. By embedding physical laws directly into the training process, PINNs enable accurate state estimation and kinetic parameter inference—even in data-limited and dynamically changing environments. Experimental results demonstrate that PINNs consistently outperform conventional data-driven models in both predictive accuracy and adaptability, while also capturing unknown or evolving system dynamics. These strengths position PINNs as highly suitable for integration into advanced control strategies such as Model Predictive Control (MPC), where reliable, real-time predictions are critical for optimizing performance, ensuring product quality, and maintaining safe, stable operation of bioprocesses.