2025 AIChE Annual Meeting
(182aw) High-Throughput Operator Engineering for Tunable Atf-Based Biosensors
Mathematical models reveal two critical equilibria—aTF-DNA binding (aTF+DNA) and aTF-effector binding (aTF+effector)—governing biosensor sensitivity. Adjusting the dissociation constant (Kd) between aTF and DNA or effector enables tuning of the half-maximal inhibitory concentration (I50) and the maximal slope of the dissociation curve (kmax). However, engineering TF mutants with optimal Kd values typically requires complex protein modifications and labor-intensive screening. In contrast, operator sequence engineering offers a more streamlined approach by modifying the DNA sequences that TFs bind to, allowing for fine-tuning of sensitivity, specificity, and dynamic range without the complexities of protein engineering.
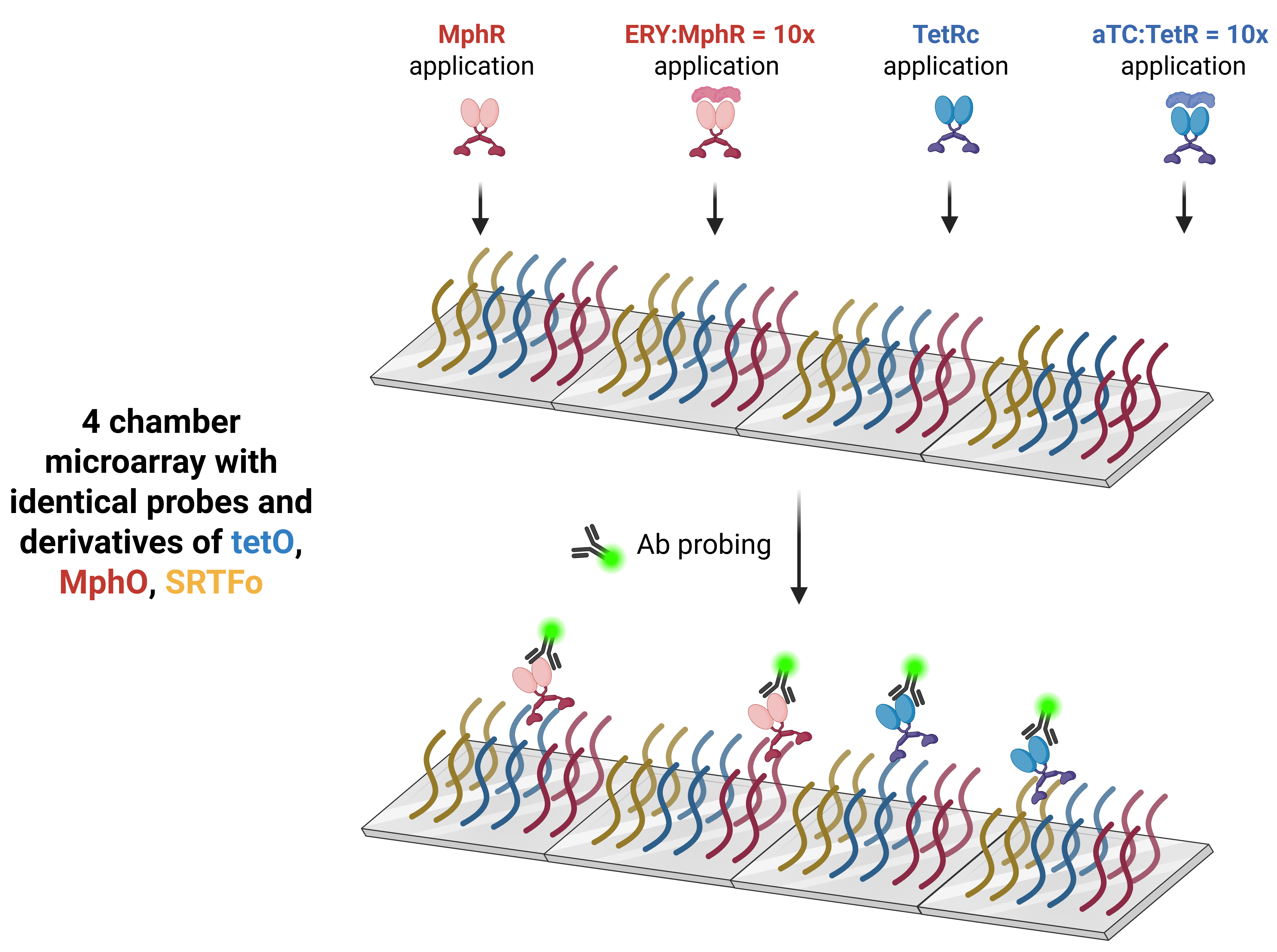
To systematically evaluate operator sequences, we repurposed protein binding microarrays (PBMs)—a high-throughput platform for in vitro characterization of DNA-TF interactions. Agilent microarrays were functionalized with operator variants containing 1–2 base pair mutations within two binding sites, generating symmetrical and asymmetrical dyads. Each microarray contained four identical chambers with immobilized DNA probes. ssDNA in triplicates of the forward/reverse orientations were double-stranded via on-chip PCR while fluorophore-conjugated antibodies detected epitope-tagged TFs bound to DNA.
DNA Sequence Conservation Analysis revealed several important takeaways. Position weight matrices (PWMs) and k-mer analysis of Chambers 1 (MphR) and 3 (TetR) identified conserved nucleotides and motifs critical for binding. Ladder plots comparing fluorescence intensity between TF-only (Chambers 1/3) and TF+ligand (Chambers 2/4) chambers revealed significant binding reduction upon ligand addition. Variants with weakened Kd values showed steeper dissociation slopes, potentially enabling tunable sensitivity. Deeper understanding of the binding affinities for the operator sequences that can be used in the aTF-DNA-effector molecule binding schemes will be used to extend the dynamic range of bead-based optical sensors for the rapid readouts of multiplexed analyte detection across wide concentration ranges.