2025 AIChE Annual Meeting
(59c) Evaluating Implicit Versus Explicit Solvent Models in Glycan Simulations
So far, there are advanced computational tools that can provide insights into the structural dynamics of complex sugars, however, studies have primarily focused sparsely glycosylated systems. Glycans are often large and structurally intricate, particularly in systems such as mucins, which are heavily glycosylated, and in HIV Envelope Protein, where the envelope is covered by a dense glycan layer. Other important biological areas, such as immune recognition and cancer progression, also involve complex glycan structures. Simulating these large glycan systems at an atomistic level is especially challenging due to the inclusion of huge number of explicit water molecules, which makes the already large system even more complex.
While an accurate solvent model is necessary to analyze water effect on large, understudied glycan systems, simulating large systems with explicit solvation remains challenging. So, we aim to understand glycan performance using implicit solvent model which does not include water molecules but represents it as a continuum medium. In this study, we modeled five glycans commonly found in the mucin glycoproteins, a protein known to be 50–80% glycosylated, and interacting with water to form hydrogels with several important physiological roles in health and disease.
In this study, we compare the effects of implicit versus explicit solvent models on glycan simulations by analyzing key structural, and thermodynamic properties. We constructed four model systems for each glycan type to study the effects of water on structural parameters: (1) a single glycan attached to a short peptide (AATAA), (2) multiple glycans attached to a peptide to capture glycan-glycan interactions, (3) a system containing charged and neutral glycans below the saturation threshold; and (4) a system containing charged and neutral glycans above saturation threshold to examine solvation effects. The distribution of root mean square fluctuation for flexibility, glycan dihedral distributions, boat-chair puckering conformational transitions, persistence length and aggregation propensity of the glycans in different water models will be presented.
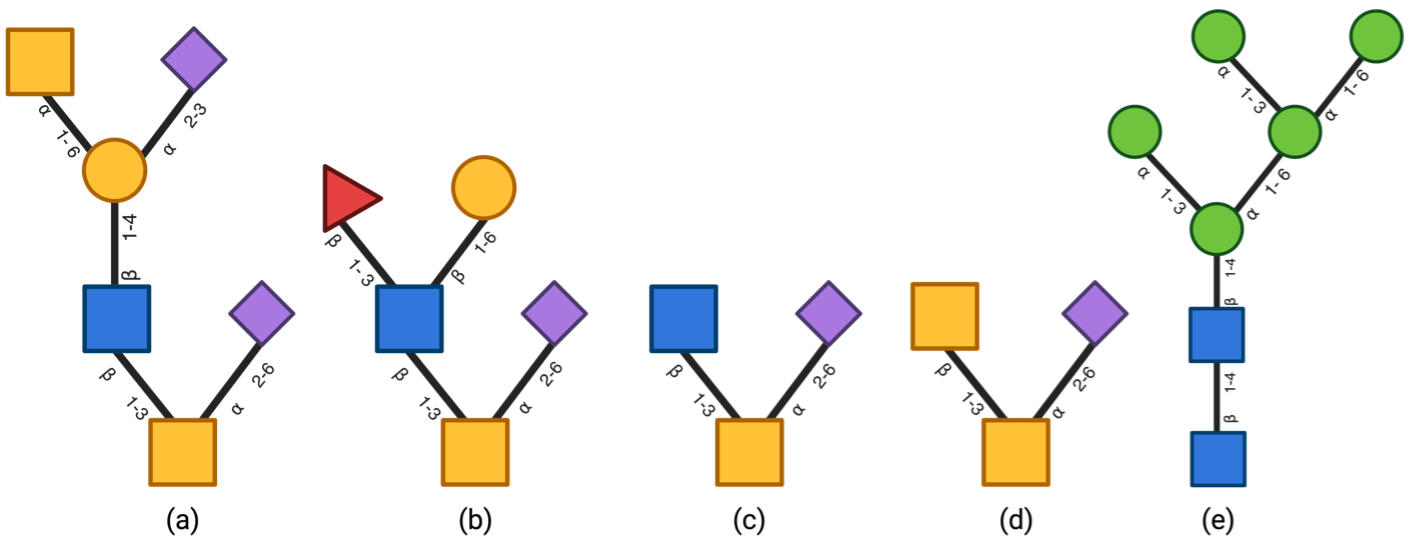
The initial structure of the glycans is modeled using CHARMM-GUI. The sequence used for our case study are shown in Fig. 1. Each system is solvated in an octahedral box with 15 Å distance between the solute and the edge of the simulation box. To compare water models, all simulations are performed using NAMD, with both an implicit (Generalized born) and an explicit (TIP3) solvent model.
To improve implicit solvent models, we explore tuning parameters of dielectric constant, SASA-based nonpolar solvation terms, and generalized Born radii to better capture glycan-specific hydration effects and conformational behavior. Our goal is to enhance the feasibility of using implicit solvent models for large, densely glycosylated systems like mucin glycoprotein. Conducting computational studies over longer timescales would be efficient, not only for mucin studies but also for other large and densely glycosylated systems which influence health and disease.
Fig .1. Commonly found glycans in mucin glycoprotein (a) [NeuAc-] GalNAc-Gal-GlcNAc-3(NeuAc-6)GalNAcol (b) SO3--Gal-(Fuc)GlcNAc-3(NeuAc-6)GalNAcol (c) GlcNAc- 3(NeuAc-6)GalNAcol (d) GalNAc-3(NeuAc6)GalNAcol (e) Man5GlcNAc2