2025 AIChE Annual Meeting
(588bv) Dynamic Modelling of Membrane-Enhanced Liquid-Phase Oligonucleotide Synthesis with Homostars
No mathematical models are currently available to describe LPOS process with homostars. In this work, we develop a dynamic model of LPOS to capture the interplay between key operational decisions and their impact on the yield and purity of oligonucleotides. Our model describes the multi-step LPOS process, where the chains attached to the homostar undergo successive reaction and diafiltration phases. Each repeated process step includes deprotection, diafiltration, coupling and a second diafiltration stage. We account for the formation of error sequences during the coupling where growing chains react with nucleotides, directly affecting product yield and purity. Our model combines reaction kinetics and membrane transport within a multistage dynamic structure.
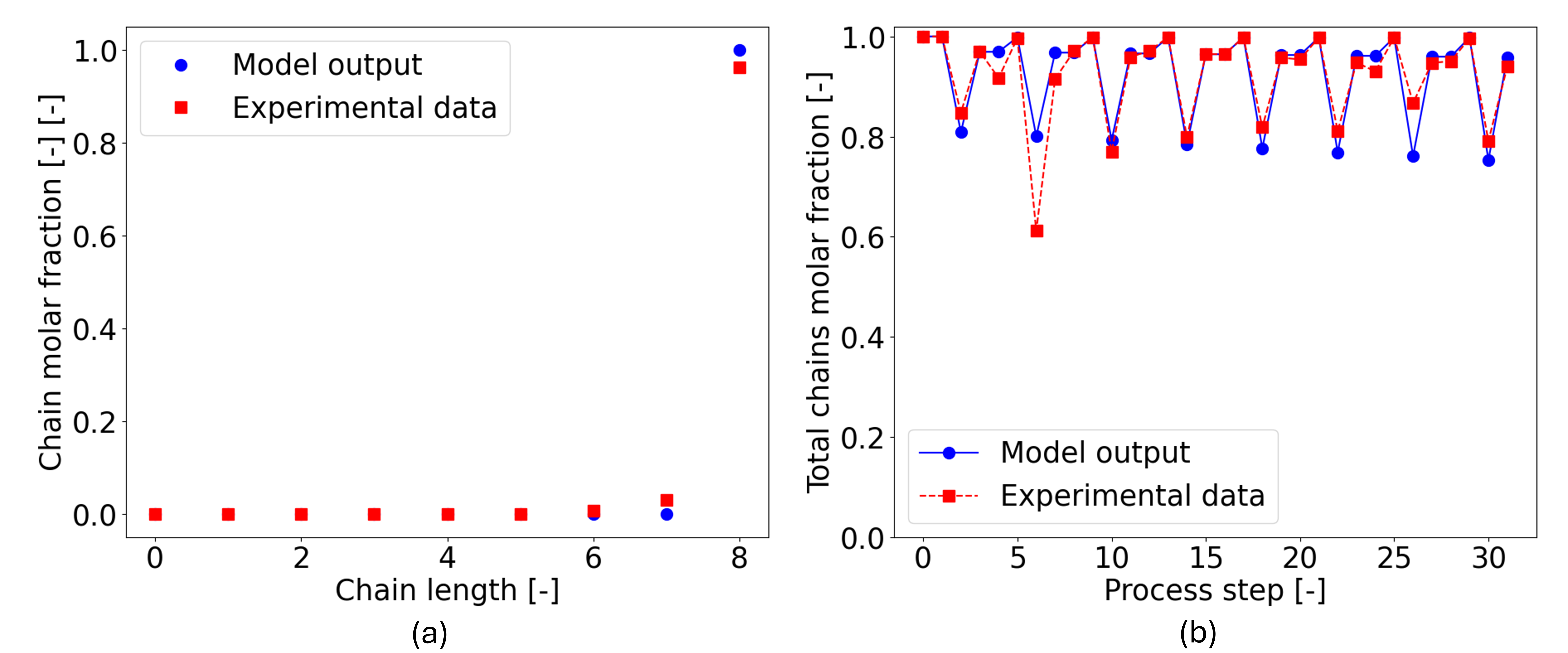
We calibrate the model against experimental data manually extracted from chromatographic elution profiles (Figure 1). The model shows good performance in simulating both the relative molar fractions of chains of different lengths at the end of the process (Figure 1a), as well as the dynamic profile of anchored chains and nucleotides molar fractions thorough the process (Figure 1b).
This calibration provides a first reliable set of estimated parameters, which we further refine using full spectral data and chromatographic elution profiles for parameter estimation, instead of relying on manually extracted data. To achieve this, we directly integrate spectral data into the LPOS model, eliminating the need for manual data extraction. We draw from the approach proposed in Chen et al. (2016) to expand the kinetic ODE model with a data-driven component to infer information on the hidden chemical composition of the system. The matrix containing spectral data is decomposed to retrieve a matrix of concentration of the chemical species and a matrix representing the individual contribution of each species to the absorption spectra (Chen et al., 2016). This bilinear decomposition is coupled with the kinetic model and all the parameters of this extended model are simultaneously re-estimated to overcome the ambiguity issues inherent in Multivariate Curve Resolution techniques (Chen et al., 2016).
Our model thus enables accurate monitoring of LPOS and can be exploited for process optimization, design space characterization, and online control strategies. This provides a useful tool for the efficient implementation of LPOS and acceleration of scalable, sustainable, and efficient oligonucleotide manufacturing.
Acknowledgements
We gratefully acknowledge Exactmer Ltd. for providing experimental data employed for model calibration.
References
Chen, W., Biegler, L. T., & Muñoz, S. G. (2016). An approach for simultaneous estimation of reaction kinetics and curve resolution from process and spectral data. Journal of Chemometrics, 30(9), 506–522.
Ferrazzano, L., Corbisiero, D., Tolomelli, A., and Cabri, W. (2023). From green innovations in oligopeptide to oligonucleotide sustainable synthesis: differences and synergies in TIDES chemistry. Green Chemistry, 25(4), 1217–1236.
Gaffney, P.R.J., Kim, J.F., Valtcheva, I.B., Williams, G.D., Anson, M.S., Buswell, A.M., and Livingston, A.G. (2015). Liquid phase synthesis of 2’-methyl-RNA on a homostar support through organic-solvent nanofiltration. Chemistry – A European Journal, 21(26), 9535–9543.
Kim, J.F., Gaffney, P.R.J., Valtcheva, I.B., Williams, G., Buswell, A.M., Anson, M.S., and Livingston, A.G. (2016). Organic solvent nanofiltration (OSN): A new technology platform for liquid-phase oligonucleotide synthesis (LPOS). Organic Process Research & Development, 20(8), 1439–1452.
Figure 1: Comparison between model simulations and experimental data for (a) the relative molar fractions of individual single-stranded oligonucleotide chains, calculated as a fraction of the total concentration of all single chains at the end of the process; and (b) the relative molar fractions of total anchored (attached) chains, calculated with respect to the combined total of attached chains and nucleotides, across all steps of the process (circles and squares represent simulated and experimental data at the end of each step).