2025 AIChE Annual Meeting
(627c) Deploying a Calibrated Population Balance Model for Further Design and Optimization of a Batch Seeded Lysozyme Crystallization Process
Authors
During the design and operation of the protein crystallization process, it is crucial to ensure critical quality attributes (CQAs) meet desired specifications. The deployment of a calibrated process model, using experimental data, can facilitate good decision-making by identifying optimal operating conditions that result in time and cost savings.
In this work, we present the development and modelling of a batch seeded precipitant-induced lysozyme crystallization process in a two-part workflow: 1) Process design by initially obtaining system understanding from unseeded experiments (NaCl as precipitant), followed by investigations into seed and impurity addition; and 2) Process understanding through calibrating a first-principles population balance model, resulting in a more optimal experimental protocol.
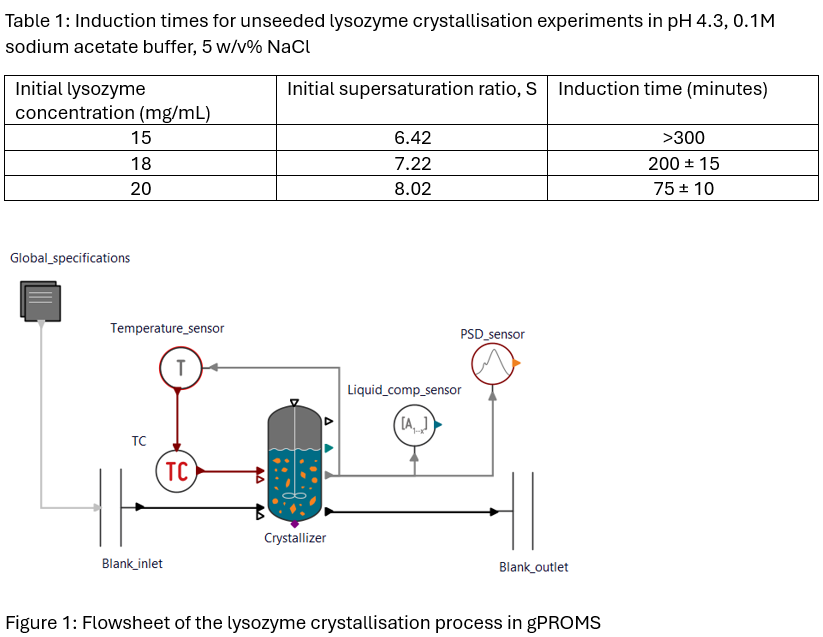
For the process design stage, 50 mL unseeded lysozyme crystallization experiments at room temperature were first conducted at varying supersaturations. These were guided by existing solubility data from literature, with the objective of obtaining an unseeded induction time of around 100-200 minutes to ensure seed-enhanced nucleation reduces induction time to a point where the process can still be adequately controlled. After an appropriate experimental protocol was obtained (initial lysozyme concentrations of around 18-19 mg/mL), silica seeds of varying attributes (porosity, particle size and pore size) were added, with the effect on PSD and induction time investigated.
To move towards process understanding, the system was then modelled using gPROMS’ Mixed Suspension Mixed Product Removal (MSMPR) Crystallizer model library, which is a first-principles process model in which necessary equations are already contained (mass, energy and population balances). Initially, conventional nucleation and growth models normally used for small molecules were selected, with gPROMS’ parameter estimation capabilities producing estimates for kinetic parameters from experimental data. Similar to previous studies investigating the applicability of conventional kinetic models to lysozyme crystallization, ln(A) values of 25-50 and surface energies of 1 mJ/m-2 were observed for classical nucleation theory, and growth rate constants of around 0.1-1 nm/min were observed (Mitchell et al., 2023).
To achieve a better fit, further empirical adjustments were required to adequately model the system, as protein crystallization is more complex when compared to smaller molecules, with existing frameworks such as classical nucleation theory being unable to predict protein nucleation rates. A quantitative assessment of the nucleation and growth models and adjustments were provided by gPROMS’ goodness of fit, bias, and lack of fit tests, along with further analysis (variance – covariance, and correlation matrices). Lastly, gPROMS’ proprietary global systems analysis (simulations using Sobol sampling) and optimisation algorithms (MINLP) allowed the exploration of product specifications that could be achieved given time/cost constraints.
In summary, this work aims to provide not only a case study demonstrating the successful design of a batch seeded protein crystallization process, but also the application of process modelling to enable better decision making. The implication of this work is that if successful, it could enable the eventual proposal of a workflow that could accelerate the adoption of protein crystallisation as a go to purification method in industry.
In future, factors affecting scale-up, the transferability of the workflow to another peptide/protein, and the transition to a continuous process could be investigated.
References:
Mitchell, H.M, Jovannus, D., Rosbottom, I., Link, F.J., Mitchell, N.A., Heng, J.Y.Y. (2023) Process modelling of protein crystallisation: A case study of lysozyme. Chemical Engineering Research and Design, 192, 268-279. DOI: 10.1016/j.cherd.2023.02.016.