2025 AIChE Annual Meeting
(595c) Data Augmentation Models for Machine Learning Applications in Stem Cell Bioprocessing
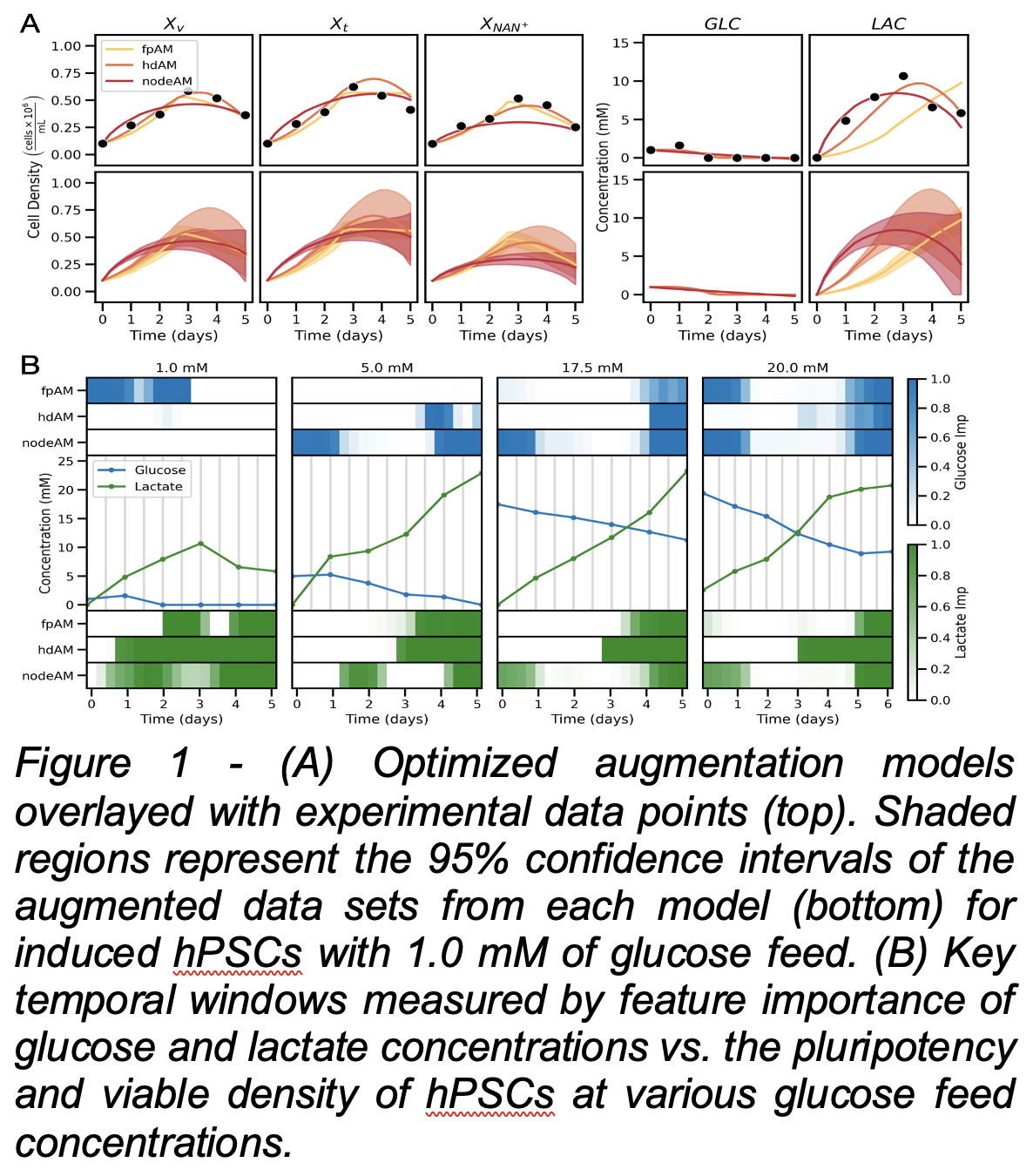
This study investigated three additive manufacturing (AM) techniques utilizing digital twins and perturbation methods to create expanded data, each varying in constraints. The first-principles AM (fpAM), which is the most constrained, relies on time-dependent material balances and physical parameters. The hybrid digital AM (hdAM) incorporates a neural ordinary differential equation (NODE) constrained by stoichiometry, growth limits, and dependencies on viable cell counts. In contrast, the statistical NODE-based AM (nodeAM) captures experimental data trends without specific physical constraints. These models amplified data for machine learning analysis of critical quality attributes (CQAs) based on glycolytic activity in hPSCs and enhanced the information represented in digital twin models. Experimental data was collected from two hPSC types (embryonic and induced pluripotent stem cells) at four glucose feed concentrations (1, 5, 17.5, 20 mM). Each AM was trained for all conditions, with representative results at 1 mM glucose displayed in Figure 1A. Feature importance concerning the relationship between glucose and lactate profiles and CQAs was assessed using a Monte Carlo variable importance projection method (Fig. 1B). Functional analysis of variance revealed that model selection significantly influences importance profiles (p = 0.014). As anticipated, lactate accumulation inhibits growth and is strongly associated with CQAs, particularly at later time points, with greater relevance observed at lower glucose concentrations. Glucose is correctly deemed unimportant when absent, and its significance increases across all models as feed concentration rises.
This study provides biologically relevant insights and enhances interpretability, addressing issues related to data scarcity. It paves the way for advancing AMs and facilitates the integration of ML in the design and optimization of bioprocesses for hPSC therapeutics and beyond.
Acknowledgment: The research was partially supported by the National Science Foundation (NSF) grant CBET-2326510.