2025 AIChE Annual Meeting
(645b) Chemeagle: An Mllm-Powered Multi-Agent System for Multimodal Chemical Information Extraction
Authors
Automated extraction methods have recently emerged as promising alternatives, primarily adopting either text-based or image-based extraction strategies. Text-based methods generally focus on recognizing chemical entities and extracting reactions from textual descriptions. Transformer-based architectures, notably fine-tuned BERT models, have significantly enhanced accuracy by identifying chemical entities and roles within reaction narratives. Meanwhile, image-based extraction involves translating graphical representations of molecules and reaction schemes into structured, machine-readable formats such as SMILES strings. Early image-based tools depended heavily on heuristic-driven algorithms, while recent approaches leverage deep learning techniques like convolutional neural networks (CNNs) and transformer-based models, notably improving robustness across various chemical drawing styles. Despite notable advancements, existing methods inherently suffer from critical limitations due to their unimodal nature. Specifically, they fail to integrate complementary contextual information dispersed across different data modalities, notably tables and supplementary graphical variants. These elements, often containing crucial details such as explicit R-group substitutions, reaction yields, and reaction conditions, have been largely overlooked by prior extraction frameworks. Recently, OpenChemIE, a rule-based framework, attempted to overcome this issue by integrating several previously developed extraction tools into a unified workflow. However, it's rule-based nature limits its scalability and flexibility. Consequently, a significant gap remains for an integrated multimodal extraction framework that systematically combines text, tables, and images to achieve comprehensive and precise chemical reaction data extraction.
Meanwhile, multimodal large language models (MLLMs) have emerged as powerful tools for information extraction tasks due to their inherent capability to integrate textual and visual information. By leveraging their cross-modal understanding and contextual reasoning abilities, these models can simultaneously interpret textual, tabular, and visual information. Such capabilities make MLLMs particularly well-suited to address the inherently multimodal nature of chemical information. A recent study, MERMaid, develped a GPT4o-based workflow that successfully integrates and aligns chemical tables with related texts, highlighting the significant potential of MLLMs in multimodal chemical extraction tasks. However, despite their potential, MLLMs still face critical limitations when applied to complex chemical data extraction tasks. While they can process text and images simultaneously, their understanding of domain-specific chemical structures and reaction mechanisms remains limited. For example, MLLMs struggle with molecular recognition and named entity recognition. Furthermore, their lack of localization capability results in poor performance in spatial association tasks, such as molecular detection and reaction image parsing. Consequently, standalone MLLMs fall short in systematically constructing accurate and structured chemical reaction datasets.
To overcome these limitations, one viable approach is to augment MLLMs with specialized extraction tools designed in the previous works. Such tool-augmented language models are commonly referred to as agents, which can autonomously invoke external computational tools to accomplish tasks beyond their inherent capabilities. A single-agent framework includes a base MLLM that integrates multiple extraction tools to sequentially manage all tasks involved in chemical data extraction, such as molecular image recognition, reaction image parsing, and named entity recognition. However, this single-agent approach often encounters significant drawbacks, such as error propagation, where inaccuracies in earlier stages negatively impact subsequent extraction tasks, and a lack of flexibility, interpretability, and robustness in handling diverse multimodal inputs. A more effective alternative is adopting a multi-agent framework, in which multiple specialized agents collaboratively manage distinct sub-tasks. Within this architecture, each agent independently focuses on a specific extraction task using dedicated computational tools. This clear specialization reduces error accumulation, enables systematic cross-validation, and significantly improves interpretability. The multi-agent approach is thus particularly suited for multimodal chemical data extraction, where crucial reaction details are scattered across text, tables, and images, necessitating precise cross-modal alignment and reasoning. By coordinating specialized agents, multi-agent systems can comprehensively extract, validate, and align multimodal chemical data, significantly improving accuracy, interpretability, and scalability.
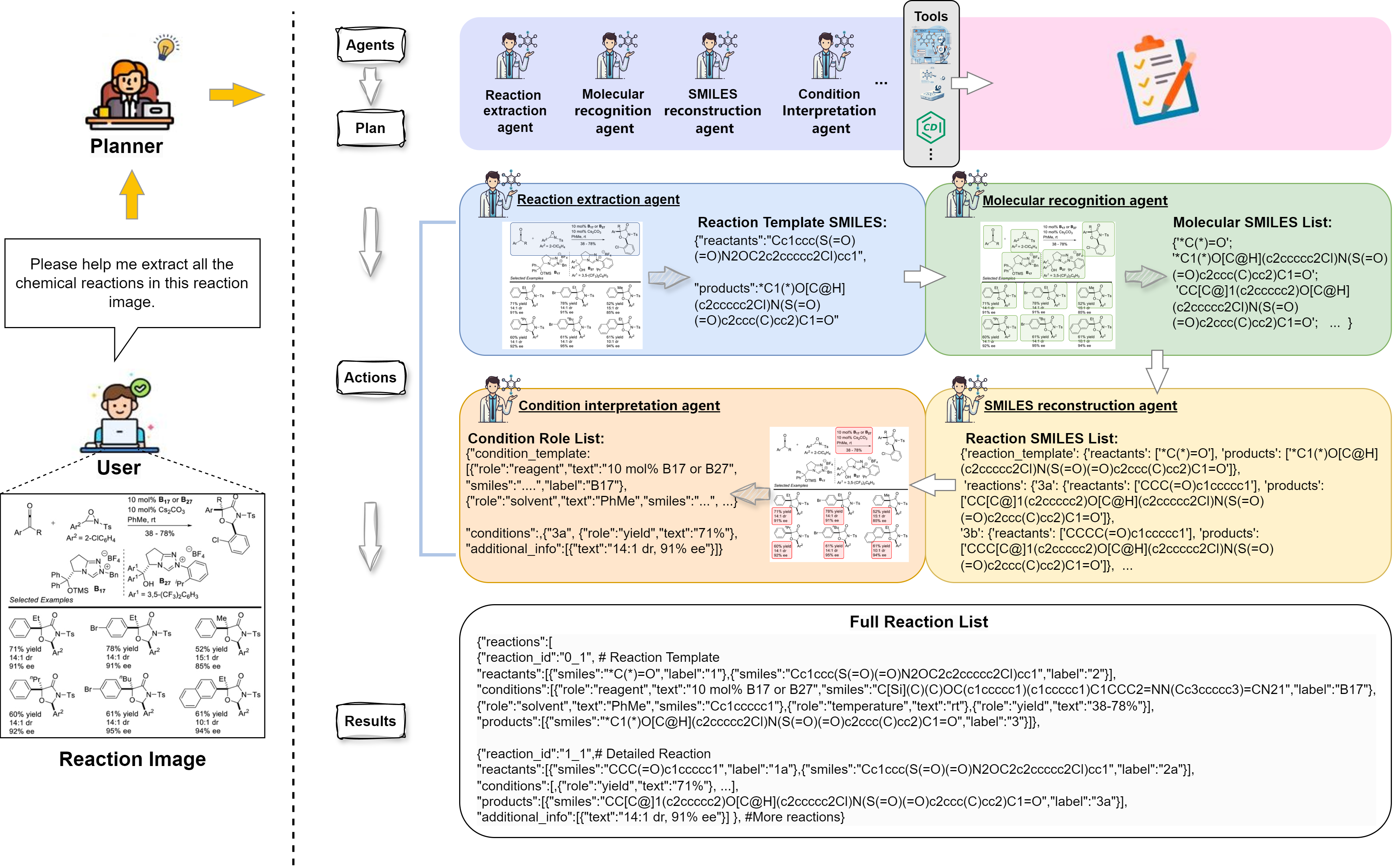
Building on the strengths of multi-agent system, we introduce ChemEagle, an MLLM-powerd multi-agent system designed to automate the extraction and integration of chemical information from multi-modal sources. ChemEagle utilizes a set of specialized agents with 10 specialized chemical information extraction tools, each optimized for a specific task such as molecular image recognition, reaction image parsing, and text-based reaction extraction. By systematically coordinating these agents, ChemEagle ensures accurate and structured data extraction across text, tables, and images. Within each agent, MLLMs still play a critical role. Specifically, the first agent called Planner leverages a MLLM to initially analyze chemical reaction images and determine an appropriate extraction workflow. The planner agent first formulates a detailed extraction strategy and sequentially assigns subtasks to the corresponding specialized agents. During subsequent extraction steps, each agent's internal MLLM continuously analyzes the original chemical images along with intermediate outputs from computational tools. This enables immediate identification and correction of common errors such as OCR errors that frequently occur in the atom sets when converting molecular structures into graph representations, which lead to inaccurate SMILES output. Additionally, the integrated reasoning capabilities of MLLMs empower agents to effectively resolve molecular coreferences, accurately align reaction conditions with their corresponding reaction schemes, and reconstruct complete substrate scopes through precise identification and substitution of explicit R-group definitions. An example ChemEagle's multi-agent workflow for extracting and structuring multimodal chemical reaction data from a reaction image with R-group annotations and a visual table of product variants is shown in our figure. ChemEagle first applies the planner agent to analyzes the reaction images and assign a series of agents for extracting chemical reactions. The process begins with the reaction extraction agent, which parses the image to identify the reaction template. The system then leverages the molecular recognition agent to detect molecular structures and generate SMILES strings for the reactants and products. The SMILES reconstruction agent subsequently refines these structures, linking them to reaction templates. Finally, the condition interpretation agent extracts reaction conditions from tables and text, associating them with the corresponding reaction components. By integrating these specialized agents, ChemEagle can process multimodal data, resolving ambiguities between text, tables, and figures. For example, reaction conditions may be referenced by labels in both the reaction diagram and a table, and ChemEagle's multi-agent system aligns these pieces of information with the correct molecular structures, yielding a complete reaction dataset. The coreference resolution capability of ChemEagle ensures that each extracted reaction component, whether from figures, tables, or text, is correctly linked to its corresponding reaction elements. By systematically integrating specialized extraction tools and MLLM-driven reasoning within a coordinated multi-agent workflow, ChemEagle achieves enhanced robustness, accuracy, interpretability, and scalability in multimodal chemical information extraction.
We rigorously evaluated ChemEagle's extraction performance on a newly constructed benchmark dataset, which includes 272 multimodal reaction diagrams with a total of 1,113 manually annotated reactions, spanning single-step and complex multi-step transformations across diverse reaction representation styles. We applied two evaluation criteria—soft-match (considering reactants and products only) and hard-match (also including reaction conditions)—to assess extraction completeness and precision. ChemEagle achieved state-of-the-art performance under both criteria, attaining F1 scores of 71.82% (soft-match) and 70.80% (hard-match). These results significantly surpassed previous multimodal frameworks, such as OpenChemIE (F1 scores of 42.95% and 35.59%) and MERMaid (F1 scores of 29.64% and 21.17%), as well as general multimodal models such as GPT-4o (9.71% soft-match, 8.48% hard-match), Qwen2.5-Max, and Llama-3.2-Vision. This superior performance demonstrates the significant benefit of ChemEagle's integrated multi-agent strategy and error correction capability. Further assessments on critical single-modal extraction tasks underscored ChemEagle's modular flexibility and robustness. In molecular image recognition tasks (evaluated on a challenging ACS dataset), ChemEagle achieved top accuracy (85.7%), clearly surpassing both specialized molecular recognition tools and general-purpose multimodal models. Similar dominant performances were observed in reaction image parsing, text-based reaction extraction, and named entity recognition tasks, validating ChemEagle's exceptional capability across individual extraction modalities.
In summary, ChemEagle offers significant advancements in automated chemical reaction extraction by successfully addressing critical limitations of previous unimodal and multimodal approaches. The innovative integration of multimodal large language model-based reasoning with specialized computational tools, orchestrated within a robust multi-agent framework, ensures comprehensive and accurate extraction across text, images, and tables. ChemEagle thus significantly improves the precision, reliability, and comprehensiveness of automatically extracted chemical reaction datasets, supporting large-scale database construction, AI-driven chemical discovery, and accelerated retrosynthesis planning. Moreover, the modular and scalable architecture of ChemEagle makes it easily adaptable, capable of incorporating emerging extraction tools and data modalities in the future. As multimodal AI and chemical informatics continue to evolve, we anticipate ChemEagles flexible framework to remain at the forefront of chemical information extraction research. Ultimately, ChemEagle's contributions pave the way towards fully automated, scalable, and accurate chemical knowledge curation, fundamentally enhancing the efficiency and effectiveness of chemical research workflows.