2025 AIChE Annual Meeting
(137h) Cell-Free Synthetic Virology for Programmable Therapeutics
Authors
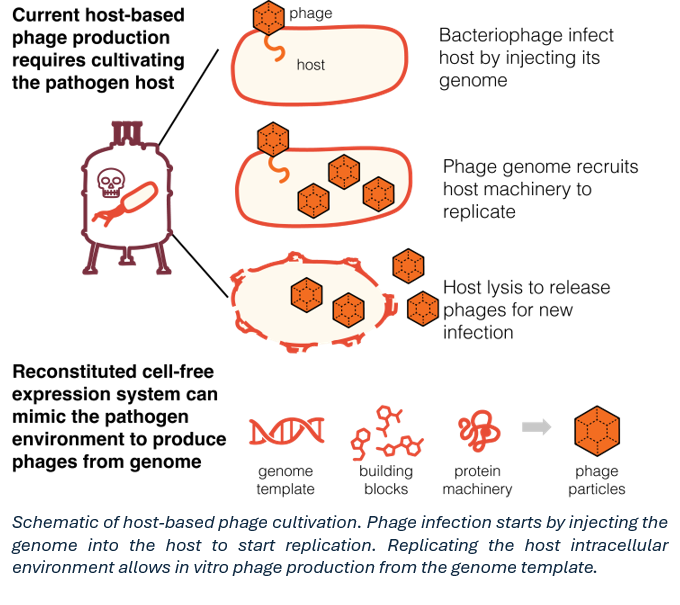
Bacteriophages – viruses that infect bacteria – offer a promising solution. They are pathogen-specific, self-amplifying, and self-limiting when the host ceases to exist. However, current methods for phage production require the cultivation of target pathogens to initiate infection and replication (Figure 1). This imposes significant biosecurity constraints and limits applicability, especially for unculturable or high-risk pathogens.
In this talk, I will illuminate my research vision in cell-free synthetic virology: a host-independent platform for bacteriophage production, engineering, and deployment to address critical challenges in antibiotic resistance and selective microbial control. Building on my past work in reconstituted cell-free gene expression systems (One-Pot PURE), I will illustrate how the same modular, “mix-and-match” workflow can be customized by adjusting transcription-translation machinery, energy regeneration schemes, and tRNA codon-usage pool to mimic various pathogen-specific intracellular environments. This approach enables host-independent synthesis of phages that are currently inaccessible through traditional methods, significantly expanding our ability to produce a broad range of bacteriophages with biomedical relevance.
Together, these efforts define a new frontier in programmable phage biomanufacturing. By decoupling phage production from live host cultivation, this approach provides a scalable, adaptive path toward combating antibiotic resistance and advancing precision microbiome engineering.