2025 AIChE Annual Meeting
(390ao) Assessing Molecular Synthetic Accessibility with Economic Indicators
Authors
To address the problem of synthesis, researchers introduced computer-assisted synthesis planning (CASP) [1], which allows the screening of synthesis pathways for a given molecule in an automated fashion. Although this is a powerful tool, CASP takes about 1 minute to compute synthesis pathways for a given molecule. However, in drug discovery routines, it is common to generate and optimize millions of molecules. Having to compute a synthesis pathway with CASP would take several years. For large-scale molecular screening, a computationally efficient alternative is needed.
To enable large-scale screening, researchers introduced "synthetic accessibility" (SA) metrics [2,3] that quantify within milliseconds whether a molecule is easy-to-synthesize (ES) or hard-to-synthesize (HS). Although existing metrics have been applied for drug discovery routines [4], they face certain shortcomings: their scoring system lacks physical interpretation and is not representative of the cost associated with molecular synthesis. Furthermore, many SA metrics are dependent on CASP to generate training data. However, CASP itself is not robust and is highly dependent on the simulation model as well as on the simulation parameters.
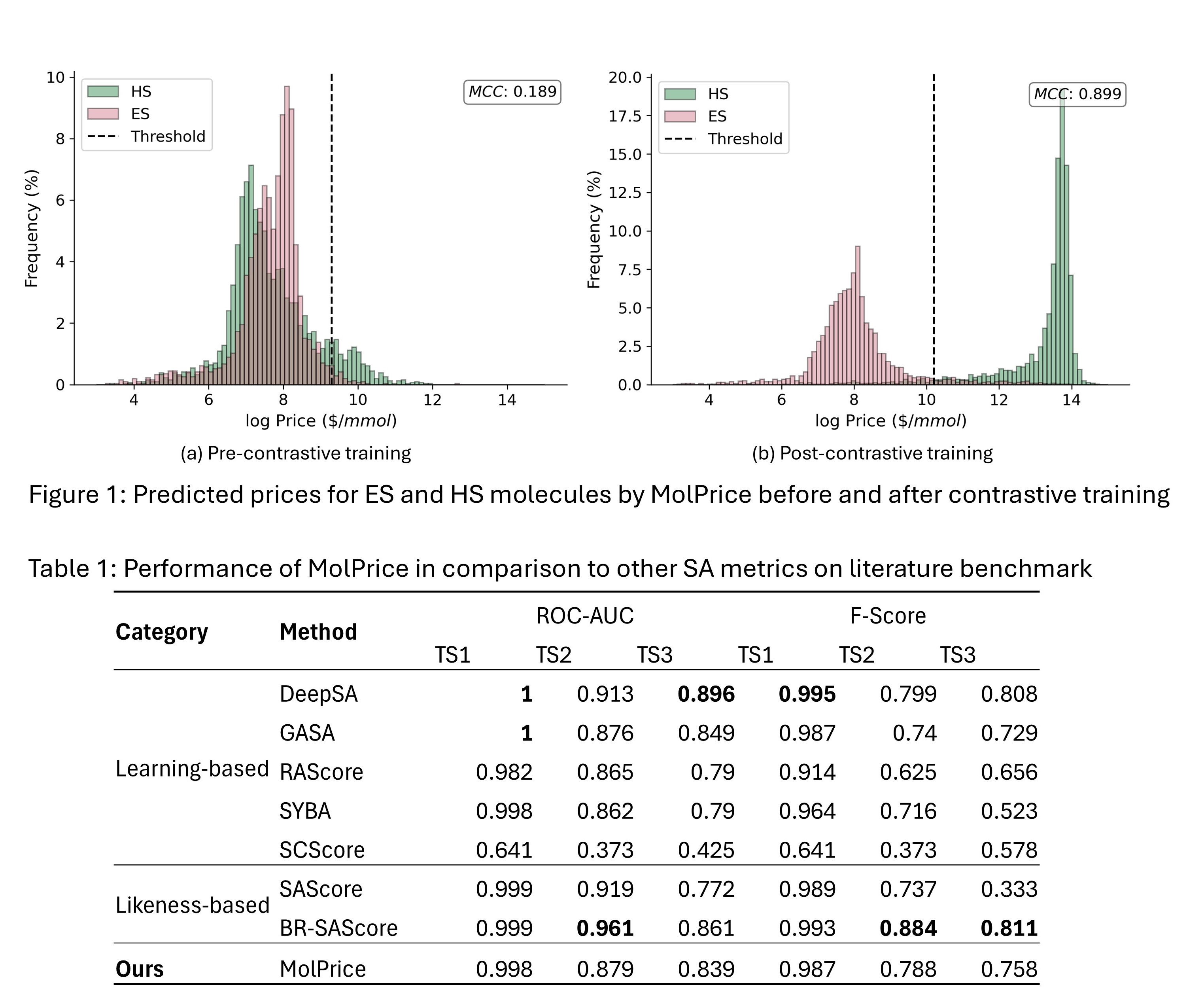
To overcome existing limitations, we propose a molecular synthetic accessibility score (MolPrice) solely based on the market price of a molecule. MolPrice is trained on an extensive dataset of 5M in-stock compounds along with their listed market price [5]. In doing so, the model learns the relationship between molecular complexity and its economic value. Because all molecules in the training dataset are "in-stock" (or generally easy-to-synthesize), we introduce a new contrastive loss based on the latent space representation of the molecule that allows MolPrice to generalize to molecules beyond its training distribution. Specifically, the contrastive loss ensures that the numerical representation of an in-stock molecule is different from that of a complex, nonpurchasable molecule. Following self-supervised contrastive learning, MolPrice reliably assigns higher prices to (unlabeled) complex molecules than to readily purchasable ones (as shown in Figure 1). Our model therefore independently learns the difference between in-stock and more complex molecules, rendering it a useful tool for SA measurement.
We compare MolPrice with existing state-of-the-art SA metrics on the standard literature benchmark [6] as shown in Table 1. Although MolPrice is not explicitly trained for synthetic accessibility performance (i.e., the classification of ES/HS molecules), it matches the performance of existing SA metrics. Furthermore, MolPrice remains highly accurate on the price prediction task with a coefficient of determination of R2=0.75. Thus, the model is capable of reliably pricing purchasable molecules while distinguishing different degrees of molecular complexity. Furthermore, MolPrice is very computationally efficient, taking only 1.3 ms to calculate the price of a given molecule. For example, the model could calculate accurate price labels for a 1M dataset in less than an hour (compared to years for CASP).
We further illustrate MolPrice's capability on a large-scale virtual screening case study. In particular, by adding MolPrice as an objective in the screening routine, molecular that are both highly purchasable and have desirable molecular properties are discovered.
In conclusion, MolPrice introduces the notion of economic value in synthetic accessibility prediction. Thanks to its computational efficiency, it can quickly screen large libraries of molecules. MolPrice bridges the gap between generative molecular design and real-world feasibility by integrating cost-awareness into synthetic accessibility assessment, making it a powerful model to accelerate molecular discovery.
References:
[1] Connor W. Coley, William H. Green, and Klavs F. Jensen. Machine Learning in Computer-Aided
Synthesis Planning. Accounts of Chemical Research, 51(5):1281–1289, 2018. ISSN 0001-4842.
doi: 10.1021/acs.accounts.8b00087.
[2] Peter Ertl and Ansgar Schuffenhauer. Estimation of synthetic accessibility score of drug-like
molecules based on molecular complexity and fragment contributions. Journal of Cheminfor-
matics, 1(1):8, 2009. ISSN 1758-2946. doi: 10.1186/1758-2946-1-8.
[3] Connor W. Coley, Luke Rogers, William H. Green, and Klavs F. Jensen. SCScore: Synthetic
complexity learned from a reaction corpus. Journal of Chemical Information and Modeling, 58
(2):252–261, 2018. ISSN 1549-9596. doi: 10.1021/acs.jcim.7b00622.
[4] Arne Schneuing, Charles Harris, Yuanqi Du, Kieran Didi, Arian Jamasb, Ilia Igashov, Weitao
Du, Carla Gomes, Tom L. Blundell, Pietro Lio, Max Welling, Michael Bronstein, and Bruno
Correia. Structure-based drug design with equivariant diffusion models. Nature Computational
Science, 4(12):899–909, Dec 2024. ISSN 2662-8457. doi: 10.1038/s43588-024-00737-x.
[5] Molport homepage. https://www.molport.com/shop/index. (Accessed: 2025-02-10).
[6] Jiahui Yu, Jike Wang, Hong Zhao, Junbo Gao, Yu Kang, Dongsheng Cao, Zhe Wang, and
Tingjun Hou. Organic compound synthetic accessibility prediction based on the graph attention
mechanism. Journal of Chemical Information and Modeling, 62(12):2973–2986, 2022. ISSN
1549-9596. doi: 10.1021/acs.jcim.2c00038.
GitHub: