2025 AIChE Annual Meeting
(151f) AI-Based Prediction of Protein Corona on DNA Nanostructures
Authors
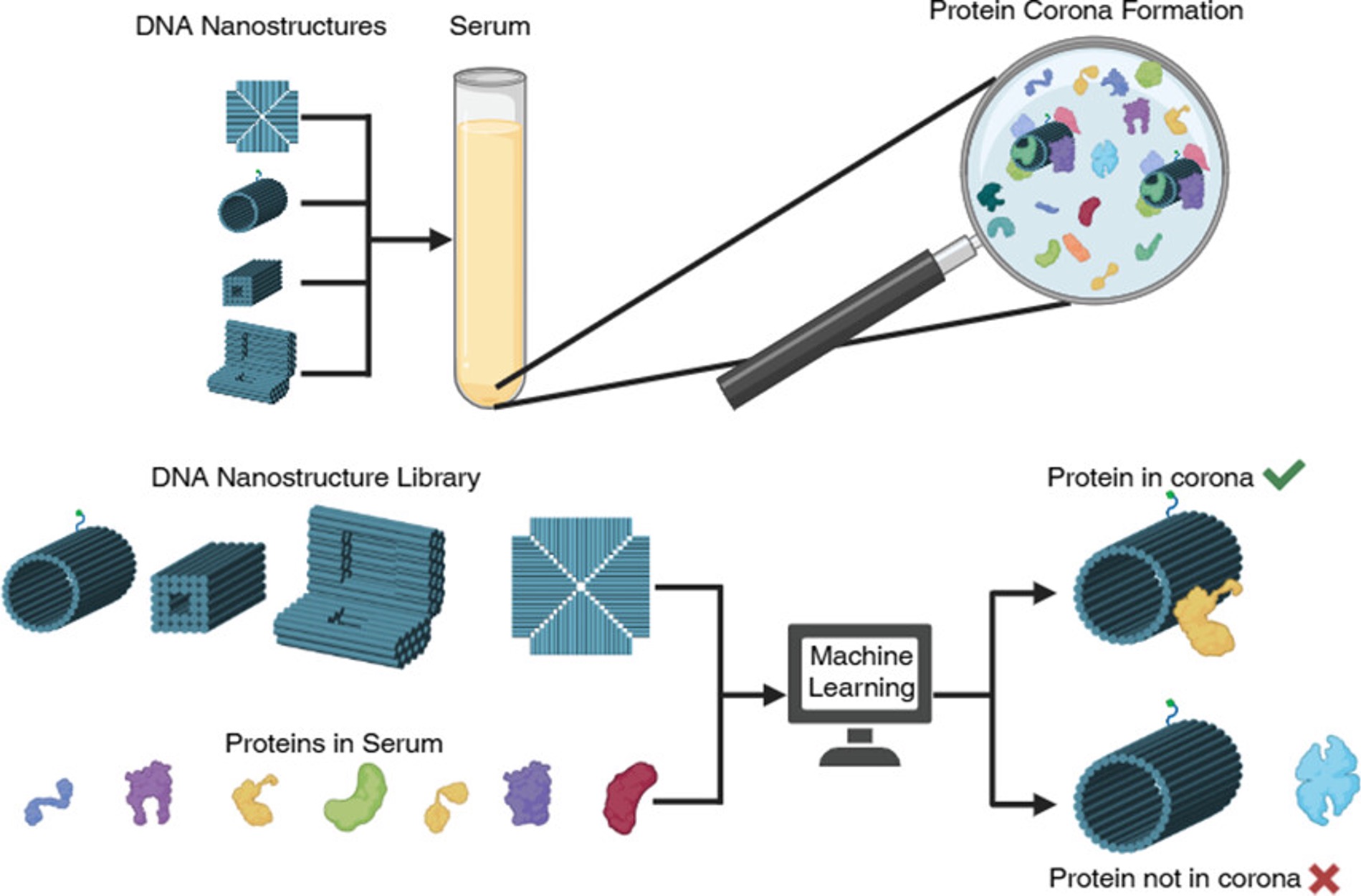
Herein, we prepared a library of 17 diverse DNA nanostructures with varying sizes, shapes, and surface chemistries to investigate the relationship between their design features and the composition of their protein corona formed in human serum. Following nanostructure incubation with the biofluid, nanostructure-corona complexes were collected through magnetic bead separation. The proteins from the nanostructure-corona complexes were analyzed with ultra-high performance liquid chromatography tandem mass spectrometry (UHPLC-MS/MS). Our proteomic results found that proteins preferentially adsorbed on distinct nanostructures mediated by their design, specifically surface modification. We then applied this proteomic dataset, including protein size, structure, amino acid composition, and biological function, along with the nanostructures’ physicochemical properties, to develop an explainable machine learning model that better elucidated the factors that impacted protein adsorption behavior on the distinct nanostructures. Our model achieved 92% accuracy in classifying protein adsorption across an array of DNA nanostructures. We also found that more than 150 features contributed to protein adsorption classification, which underscores the vast convolution of nanostructure-corona complexes. To interpret the importance values of each feature, we calculated Shadley Additive exPlanations (SHAP) values and found that the most important features that drive DNA nanostructure-corona complexes were nanostructures’ zeta-potential and non-DNA surface modifications, as well as proteins’ secondary structures and isoleucine residue exposures. We anticipate that these findings, and our first-of-its-kind nanostructure-corona predictive tool, will facilitate the development of efficacious DNA-based nanostructures for in vivo applications.