2025 AIChE Annual Meeting
(687b) Accelerating High-Concentration Monoclonal Antibody Development with Large-Scale Viscosity Data and Ensemble Deep Learning
Authors
Pin-Kuang Lai, Stevens Institute of Technology
Jia-Min Chu, Stevens Institute of Technology
Neil Mody, AstraZeneca
Jenna Caldwell, AstraZeneca
Alison Hinton, AstraZeneca
Madison Kreitz, AstraZeneca
Mitali Shah, AstraZeneca
Austin Gallegos, AstraZeneca
Valentin Stanev, AstraZeneca
Maryam Pouryahya, AstraZeneca
Mehdi Boroumand, AstraZeneca
Bismark Amofah, AstraZeneca
Amber Lee, AstraZeneca
Mark Hutchinson, AstraZeneca
Chacko Chakiath, AstraZeneca
Andrew Dippel, AstraZeneca
Gilad Kaplan, AstraZeneca
Melissa Damschroder, AstraZeneca
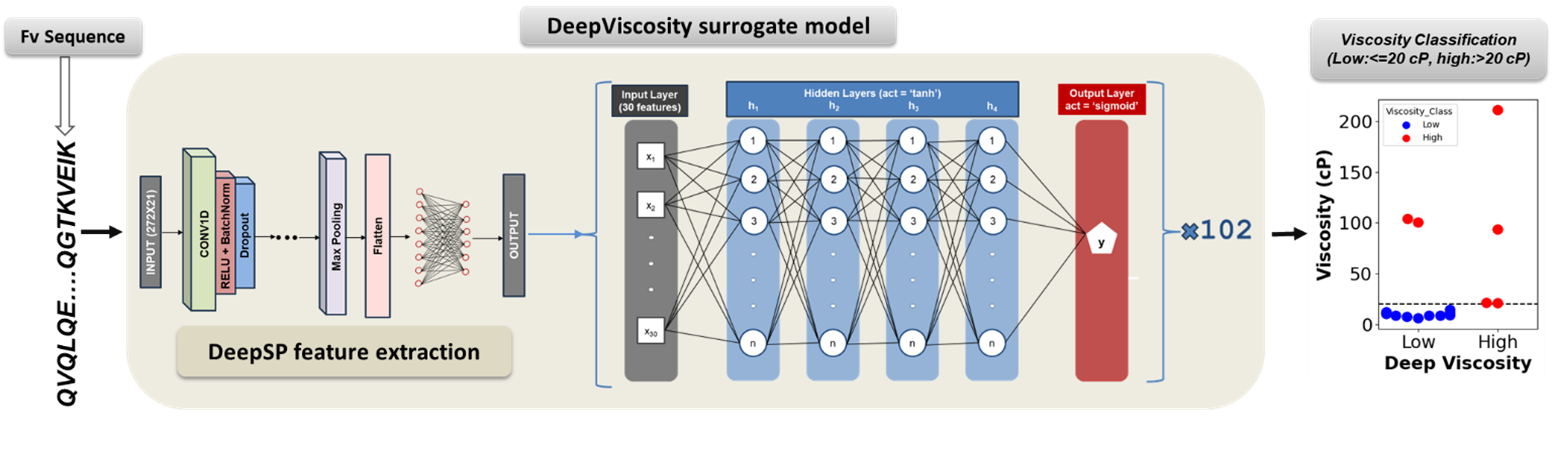
Highly concentrated monoclonal antibody (mAb) solutions are critical for subcutaneous injections but often face challenges due to high viscosities, which impact drug development, manufacturing, and administration. Existing computational models either require experimental data, structural information, computationally expensive molecular dynamics simulations; rely on complex software for feature extraction, or were trained on limited datasets of only a few dozen data points, which hinders their generalizability. In this study, we developed DeepViscosity, an ensemble model consisting of 102 artificial neural networks trained on the viscosity data of 229 high-concentration mAbs—the largest dataset recorded in this field. This model classifies mAbs as low-viscosity (≤ 20 cP) or high-viscosity (> 20 cP) at 150 mg/mL based on sequence-derived features obtained from the DeepSP model. We grouped the mAb sequences into 102 clusters using pairwise Levenshtein distance and hierarchical clustering, and employed a leave-one-group-out model validation approach. Two independent test sets (16 and 38 mAbs) with known viscosities, obtained from previous studies, were used to assess the model’s generalizability. DeepViscosity achieved training and leave-one-group-out validation accuracies of 80.7% and 88.2%, respectively, and test accuracies of 87.5% and 89.5%, outperforming other predictive models. This model facilitates early-stage mAb development by enabling the selection of low-viscosity antibodies, which is crucial for improving manufacturability and formulation properties for subcutaneous drug delivery. The DeepViscosity web-based application is freely accessible at https://devpred.onrender.com/DeepViscosity, with source codes and parameters available at https://github.com/Lailabcode/DeepViscosity. The full study is published in the mAbs journal: https://www.tandfonline.com/doi/full/10.1080/19420862.2025.2483944.