Over the past two decades, recombinant variable domains of heavy-chain only antibodies – referred to herein as nanobodies – have become an increasingly valuable diagnostic and therapeutic tool in cancer and other rare diseases. However, a significant challenge in the clinical adoption of these nanobody therapies has been the creation of stable, chemically defined, and highly specific conjugation sites within the nanobody. Stable nanobody-protein conjugates formed through genetic fusion are restricted to linkages on the N and C terminals, which can be deleterious to their binding affinity and activity
in vivo. Exploration of the nanobody-protein conjugate combinatorial design space is also limited by the cost and labor of genetic fusion, which requires the generation of an individual plasmid per new nanobody-protein combination.
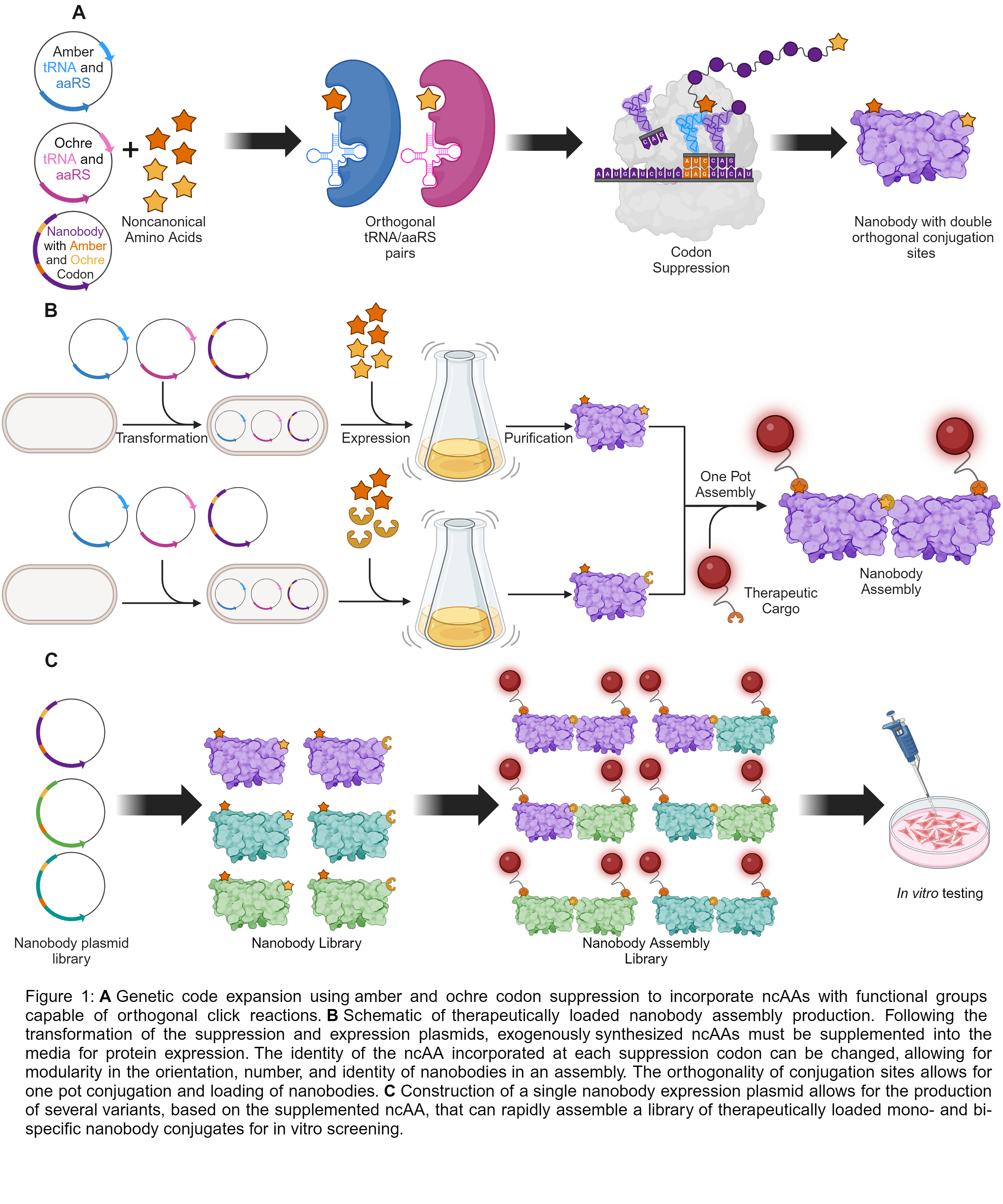
Here, we focus on the development of a platform to accelerate the generation of nanobody assembly libraries – specifically mono- and bi-specific conjugates of nanobody domains – without the need for genetic fusion to improve the retention of binding activity. Our strategy relies on: (1) the synthesis of functionalized noncanonical amino acids (ncAAs) capable of orthogonal click reactions and (2) suppression and reprogramming of stop codons for incorporation of these ncAAs into nanobodies to provide defined, stable, bioorthogonal conjugation sites. We constructed variants of immunomodulating nanobodies featuring a genetically engineered ncAA for direct incorporation into the platform. The orthogonality of encoded conjugation sites also enables this platform to be a modular, high throughput means for rapid cargo loading of nanobody assemblies, which has the versatility to include small molecules, oligonucleotides, and peptides/proteins. Analysis of glioblastoma (GMB) cell viability and receptor binding strength in vitro will allow us to evaluate the potential synergistic interactions of both agonistic and antagonistic nanobody domains and assess the structure-function relationship between the ncAA conjugation site and protein scaffold activity. Importantly, the design rules established from this work will enable the generation of a tunable system to rapidly produce synergistically active therapeutically loaded protein scaffolds for use in cancer immunotherapy.