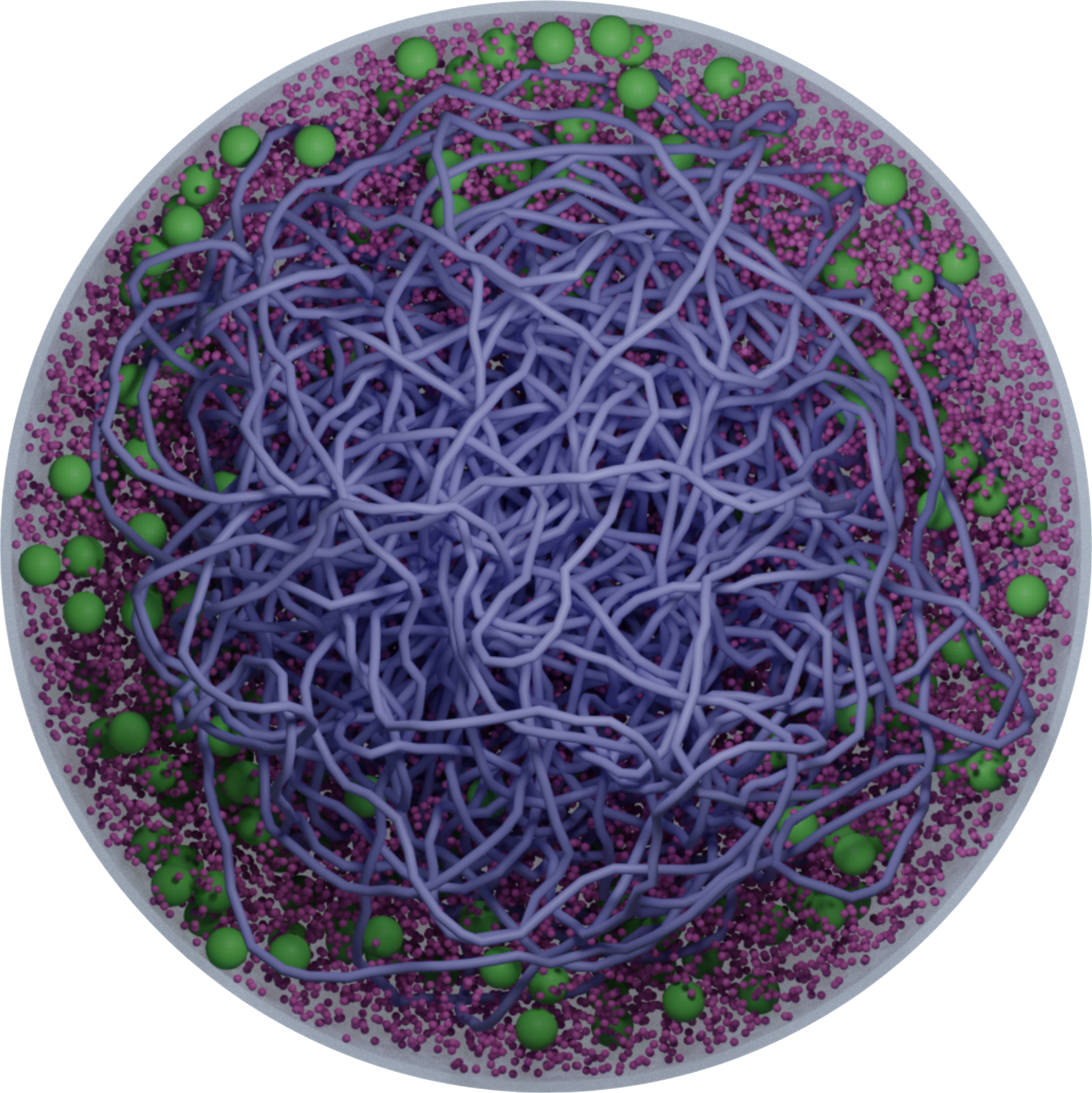
The spatial organization of the genetic material inside cells can influence intracellular processes such as transcription and translation. In bacteria such as E. coli, this genetic material is unbound but condensed into a nucleoid. This nucleoid partitions the cell into a porous region within, where transcription takes place, and a surrounding cytoplasm of mobile biomolecules where mRNA translation takes place at ribosomes. The integrity of the nucleoid results from a combination between nucleoid-associated proteins (NAPs) and osmotic pressure from cytoplasm biomolecules outside the nucleoid. In a recent work
1, we observed that the E. coli nucleoid can expand and contract, changing its density and thus promoting up- and down-regulation of protein expression by physical and electrostatic segregation of biomolecules in or out of the nucleoid. To gain further insight about the role of the latter in cell fitness, we explore the effects of such mechanisms in the JCVI-syn3A synthetic cell, which is a model bacterium comprising the smallest known genome capable of life
2,3. Syn3A offers a testbed for understanding the general principles of bacterial life. In contrast to E. coli, recent experiments
4 suggest that ribosomes in Syn3A are distributed throughout the cell rather than in an annulus near the cell wall, suggesting that the nucleoid is a loose, cell-spanning, tangled loop. In this work, we develop a coarse-grained model of the JCVI-syn3A minimal cell to capture the essential physics of DNA, native proteins, and ribosomes. Our model comprises a spherical enclosure, cytosol, thousands of explicitly resolved biomolecules, and the chromosome, which is modeled as a semi-flexible bead chain. By combining Monte Carlo simulations for chromosome generation and Brownian Dynamics, we studied the effects of native protein concentration, charge distribution, and DNA-bending HU proteins, which are modeled as a random distribution of bending defects across the genome. We find that increased concentration of HU proteins may compact the chromosome and push ribosomes out into the surrounding cytoplasm, and that changes in crowder concentration and charge distribution can alter this effect. We propose how this may be connected to the experimental observation of a cell-spanning Syn3A nucleoid.
1. Valverde-Mendez, D., Sunol, A.M., Bratton, B.P., Delarue, M., Hofmann, J.L., Sheehan, J.P., Gitai, Z., Holt, L.J., Shaevitz, J.W. and Zia, R.N., 2024. Macromolecular interactions and geometrical confinement determine the 3D diffusion of ribosome-sized particles in live Escherichia coli cells. bioRxiv, pp.2024-03.
2. Gibson, D.G., Glass, J.I., Lartigue, C., Noskov, V.N., Chuang, R.Y., Algire, M.A., Benders, G.A., Montague, M.G., Ma, L., Moodie, M.M. and Merryman, C., 2010. Creation of a bacterial cell controlled by a chemically synthesized genome. Science, 329(5987), pp.52-56.
3. Hutchison III, C.A., Chuang, R.Y., Noskov, V.N., Assad-Garcia, N., Deerinck, T.J., Ellisman, M.H., Gill, J., Kannan, K., Karas, B.J., Ma, L. and Pelletier, J.F., 2016. Design and synthesis of a minimal bacterial genome. Science, 351(6280), p.aad6253.
4. Gilbert, B.R., Thornburg, Z.R., Lam, V., Rashid, F.Z.M., Glass, J.I., Villa, E., Dame, R.T. and Luthey-Schulten, Z., 2021. Generating chromosome geometries in a minimal cell from cryo-electron tomograms and chromosome conformation capture maps. Frontiers in Molecular Biosciences, 8, p.644133.