Therapeutic antibodies have received widespread interest and investment in recent years due to their ability to treat a variety of conditions including cancer, autoimmune diseases, and viral infections. Developing high-affinity antibodies often includes many laborious and time-consuming molecular cloning steps. Surface display using
Saccharomyces cerevisiae is frequently employed to screen antibody candidates, but a diversified library must first be generated and transferred into yeast. To address this limitation, we are building a yeast-based platform that is capable of generating antibody diversity by mimicking how human antibodies are refined in B cells â by altering DNA sequences through somatic hypermutation. We have developed a method to recapitulate somatic hypermutation in yeast
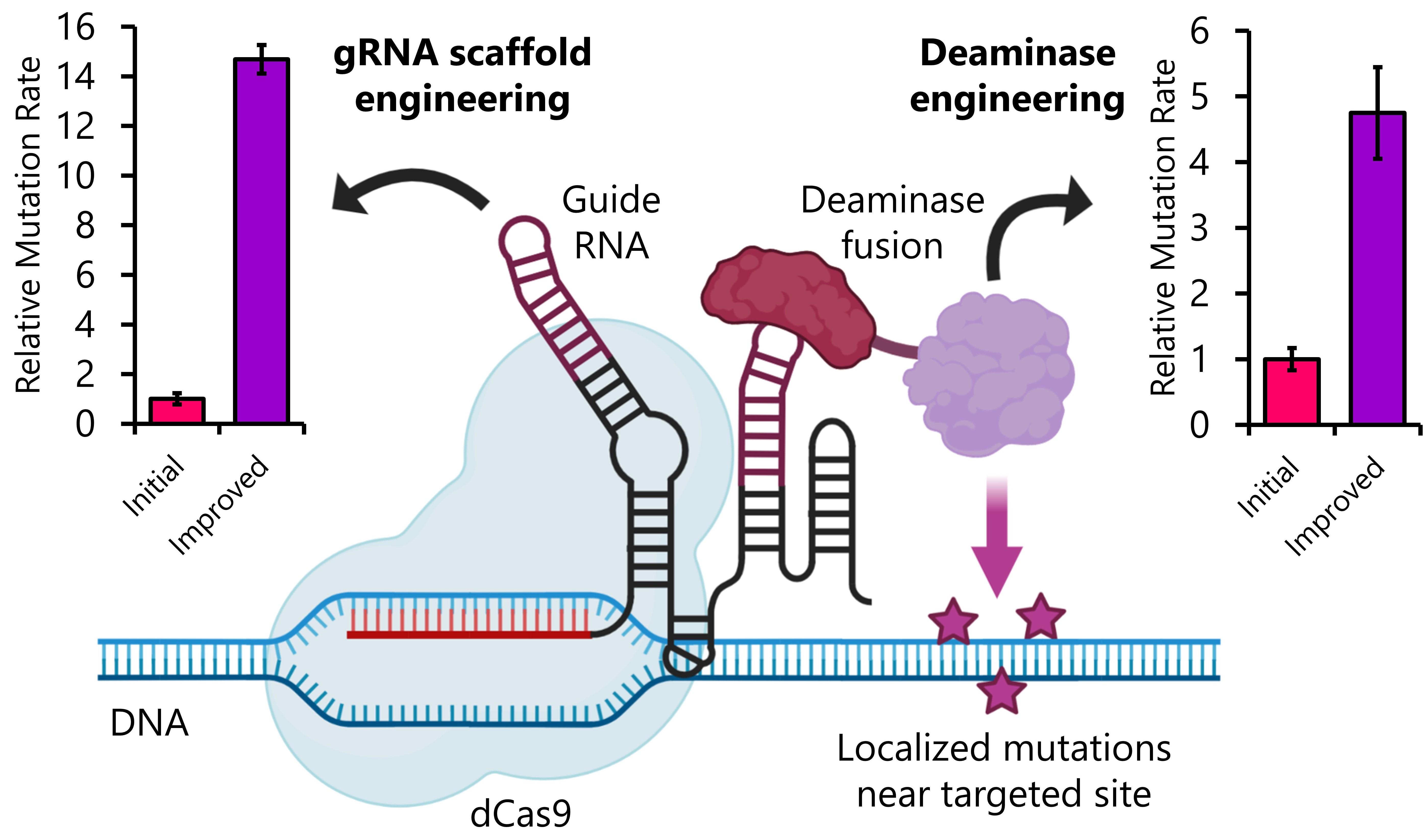
in situ using a CRISPR-based, âdiversifying base editorâ (DBE). Our system creates intense, localized mutations on a targeted gene (e.g., an antibody sequence) while minimizing off-target effects. Our DBE consists of a nuclease-dead Cas9 protein that can be complexed with multiple human deaminase protein fusions recruited via guide RNA scaffold sequences, allowed precise mutation of a targeted locus (
Figure 1).
We will highlight three key results from our efforts: (1) Fluorescence-based, mutational assays that have allowed us to estimate the rate and breadth of mutations produced by the DBE, (2) Protein engineering efforts that enhanced the mutation rate of the DBE 5-fold, and (3) Optimization of the guide RNA scaffold sequences to further enhance mutation rate up to 15-fold (Figure 1). Finally, we will describe ongoing experiments employing our optimized DBE and yeast display for the in situ enhancement of antibody affinity and isolation of these sequences in a manner that mimics B cell somatic hypermutation. Ultimately, we foresee our system becoming a rapid, inexpensive, and easily implemented tool for antibody therapeutic development pipelines, as antibodies can be improved by applying multiple rounds of DBE-mediated mutagenesis and FACS selection.
Figure 1. A CRISPR diversifying base editing (DBE) causes localized mutations in a targeted gene. The DBE is composed of a nuclease dead Cas9 (dCas9), a deaminase fusion, and a guide RNA with aptamers. By engineering the guide RNA and deaminase, we have increased the mutation rate of the DBE.