Breadcrumb
- Home
- Publications
- Proceedings
- 2022 Annual Meeting
- Food, Pharmaceutical & Bioengineering Division
- Engineering Protein Therapeutics
- (21b) Engineering Pan-Reactive VEGF Antagonists for Neovascular Eye Diseases

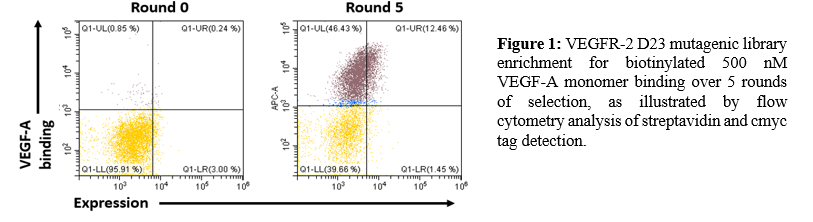
Materials and Methods: VEGF-A and VEGF-C dimers were produced in a mammalian cell expression system and biotinylated for use as targets in library selections and titrations. We cloned an error-prone mutagenic library of VEGFR-2 into the pCT yeast expression plasmid and transformed the resulting DNA into electrocompetent EBY-100 yeast. We performed five rounds of magnetic- and fluorescence-activated cell sorting (MACS and FACS) against VEGF-A to select for functionally- expressing, high-affinity variants. Variants that bound VEGF-A were then tested for VEGF-C binding. Sequences were extracted from clones that bound both ligands, and the evolved VEGFR-2 D23 mutants were titrated against VEGF-A and VEGF-C in order to determine their affinities for each ligand.
Results and Discussion: VEGFR-2 D23 was found to display on the surface of yeast cells when plasmid DNA encoding the protein sequence with a C-terminal cmyc epitope tag was transformed into competent yeast, but binding was unable to be detected for either VEGF-A or VEGF-C when the biotinylated ligands were incubated with fluorescent streptavidin. An error-prone library with a mutation rate of 1.9 amino acids/variant was cloned and transformed into yeast. We demonstrated the enrichment of the mutagenic library against VEGF-A over 5 rounds of selections (Figure 1), and were able to identify several variants with a range of affinities that were able to bind both VEGF-A and VEGF-C with high affinity.
Conclusions: We have identified and characterized high-affinity VEGFR-2 D23 variants that engage VEGF-A and VEGF-C. These clones are being further characterized and will be the basis for a site-directed library based on the crystal structures of VEGFR-2 interacting with VEGF-A and VEGF-C aimed at further increasing the affinity of the antagonist for both ligands.