2022 Annual Meeting
(19e) Generating Extended Grnas with Hairpins for Increased CRISPR Editing Specificity
Authors
Hillary Dimig - Presenter, North Carolina State University
Eric Josephs, Duke University
Ashley Herring-Nicholas, University of North Carolina at Greensboro
Miranda Roesing, University of North Carolina at Greensboro
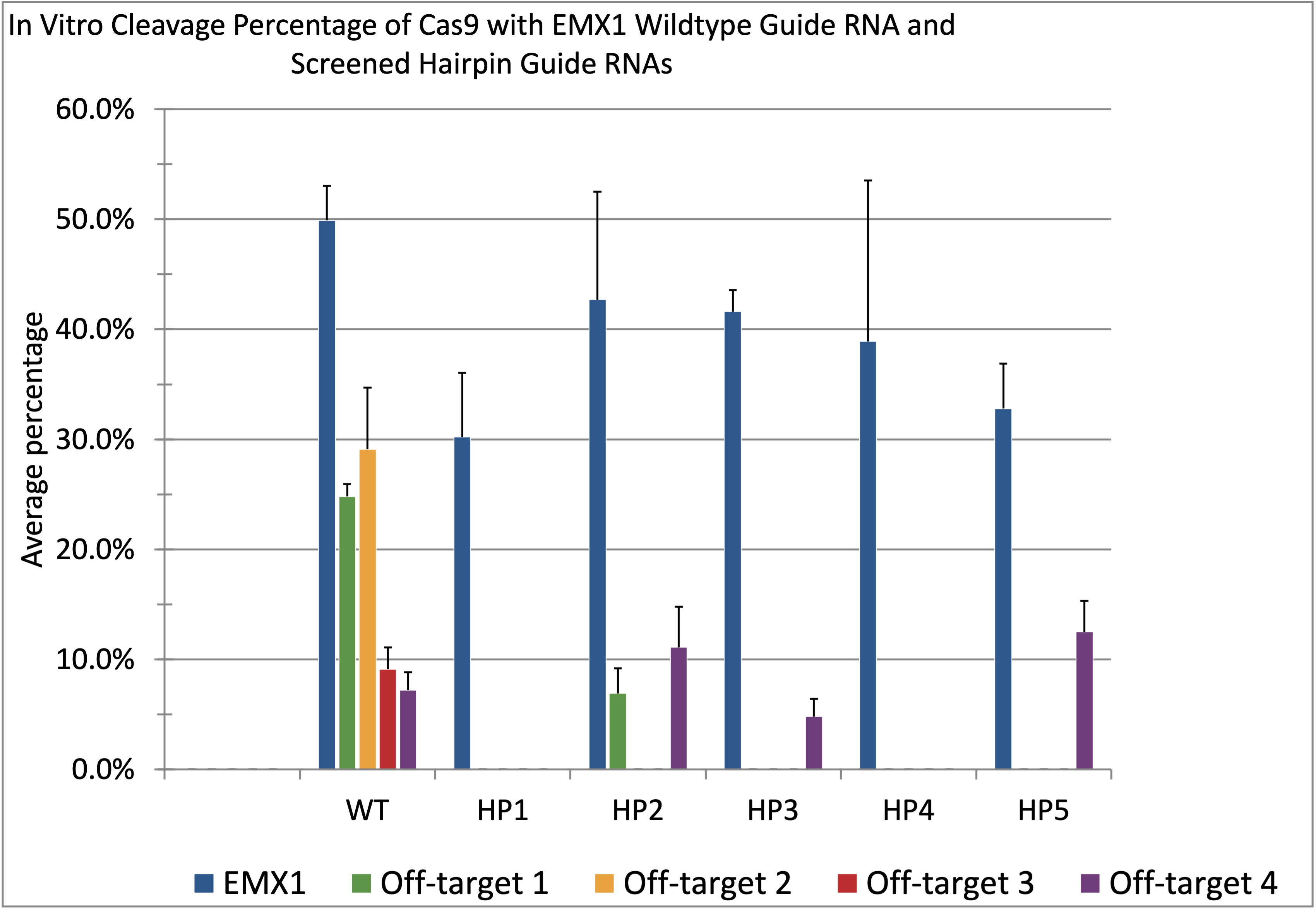
CRISPR, or clustered regularly interspaced short palindromic repeats, systems have gained notoriety for their gene editing ability. Clinical trials using CRISPR systems are currently underway for several blood diseases that are difficult to treat otherwise including sickle cell disease, inherited eye disease, and other conditions. However, off-target editing is still a concern. For example, for EMX1, a well-characterized clinically relevant gene, on-target editing accounted for 4,521 events, while the most common off-target accounted for 3,123 editing events. Extensions of the 5â sgRNA with secondary structure have been shown to increase specificity by on average 55-fold, but there is no good method for designing these extensions. We developed a method for generating guide RNAs with 5â extensions that result in high on-target efficiency and low off-target editing. For EMX1, this method was used to generate sgRNAs that were able to reduce off-target cleavage at the top four most common off-targets while maintaining on-target cleavage efficiency of the wild type sgRNA. In most cases no cleavage was observed at these off-target sites.