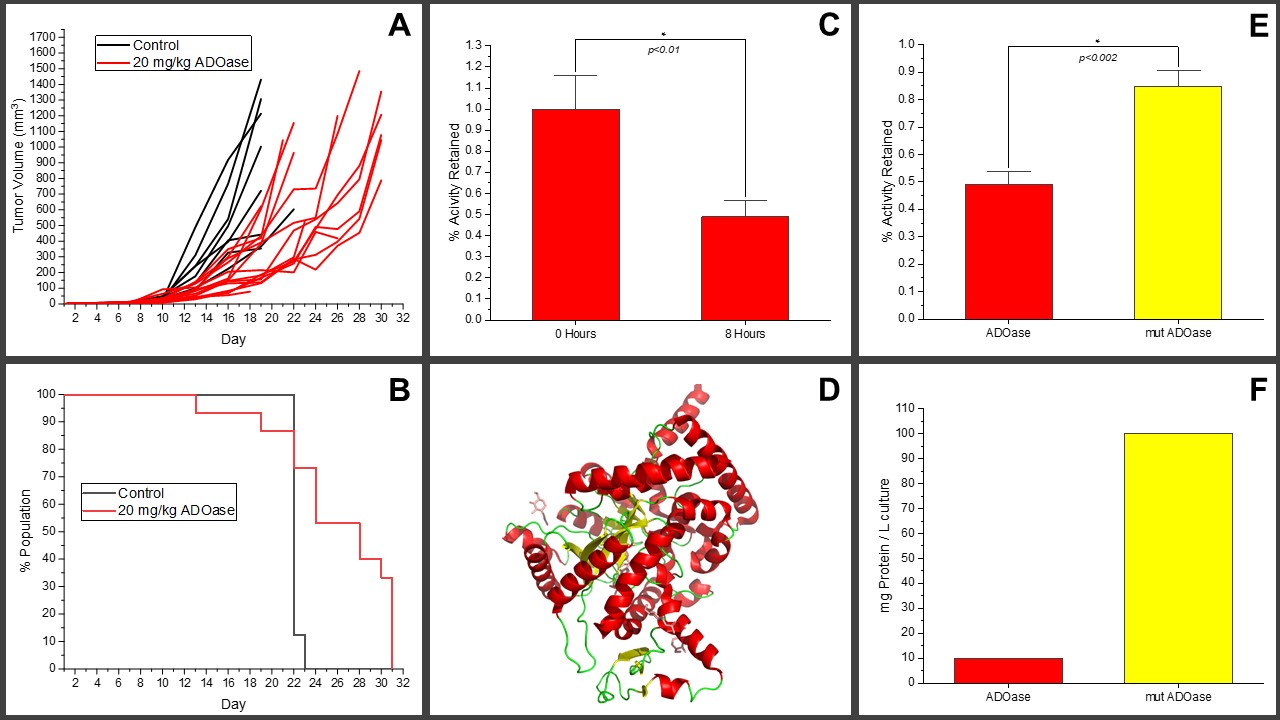
Adenosine serves important functions in the body such as inducing sleep and calming immune cells to allow healing in damaged tissues. When cancer cells in tumors upregulate the machinery to produce adenosine, local concentrations of the molecule can reach a 1000-fold increase relative to normal tissue. Such high adenosine concentrations deactivate immune cells that may otherwise recognize and destroy the cancerous cells. An adenosine-degrading enzyme, ADOase, breaks down adenosine with a high catalytic efficiency on the order of 10
6 M
-1s
-1. We have shown that administration of ADOase to a colorectal cancer mouse model slowed tumor growth (Fig 1A) and delayed mortality significantly (Fig 1B). However, ADOase loses ~50% of its activity within eight hours at physiological conditions (Fig 1C). We solved the crystal structure of our enzyme (Fig 1D), which allowed us to identify structural motifs that could contribute to its instability through comparison to enzymes that do not deactivate at physiological conditions. Structural and sequence alignment of ADOase with similar enzymes demonstrated regions conserved amongst more resilient proteins that differed from the associated ADOase sequences. We thus rationally determined particular amino acid positions within ADOaseâs makeup for mutagenesis that could potentially curb the enzymeâs tendency to deactivate. To streamline and hasten evaluation of each variant for activity retention, we developed an
Escherichia coli lysate-based assay to allow characterization of the stability of enzyme variants in a more high throughput manner, allowing us to isolate variants with improved stability after 8 hours (up to ~90% activity retained) at physiological conditions (Fig 1E), and serendipitously, with 10-fold improved yield retained from affinity chromatography purifications relative to the wild-type enzyme (Fig 1F). Armed with a mutant form of ADOase less likely to degrade at physiological conditions, we now plan to determine the therapeutic efficacy and pharmacokinetic/pharmacodynamic profile of the mutant compared to that of the original enzyme in tumor mouse models. In summary, we detail an engineering workflow to identify enzyme mutants with reduced tendency to deactivate at a given temperature that can be customized to meet the needs of other protein engineers.